CombiRIDGE - High Throughput Combinatorial Anchored Docking for Virtual Screening of Vast Chemical Spaces |
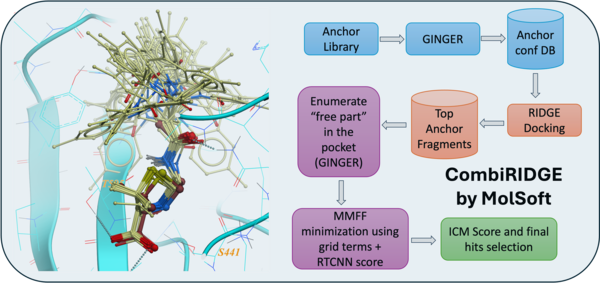
CombiRIDGE is a new innovative GPU-accelerated solution for high-throughput ligand docking and screening which leverages MolSoft's generative neural network conformer enumeration method GINGER, ultra-fast GPU docking technology RIDGE and advanced graph neural network scoring ( RTCNN ). The approach allows you to optimize specific R-groups and screen vast ultra-large or combinatorial libraries efficiently.
The anchor-driven approach begins with a 3D anchor or core fragment and a 2D R-group, anchoring the core within the binding site to maintain critical interactions. Using GINGER, full 3D conformers are generated directly at the binding site, allowing very rapid combinatorial expansion of pre-docked anchor structures into complete compounds that sample the full library space around each anchor fragment. Pose refinement is achieved through GPU-accelerated grid/MMFF energy minimization, optimizing poses for enhanced accuracy and consistency. Finally, advanced scoring using RTCNN evaluates docked poses, providing robust metrics for selecting high-quality candidates.
An Anchor Library is a curated collection of small molecules specifically designed to efficiently explore the chemical space surrounding a particular molecular scaffold. MolSoft has curated the following Anchor Libraries from three leading vendors, with more to come.
| Vendor | Anchor Library Size | Total Space |
|---|---|---|
| Enamine REAL | ~45M | ~48B |
| Wuxi Galaxi | ~35M | ~15B |
| eMolecules | ~60M | ~1.2T (>7T soon) |
In this example, 28 billion chemicals from the Enamine REAL database were screened using a single Nvidia RTX 4090 GPU.
The process begins with anchor pre-processing using GINGER. Approximately 18 million anchor molecules were converted into 3D conformers at a rate of 7,500 per minute, completing the task within 40 hours.
Next, anchor docking was performed with RIDGE. Small anchor molecules with low rotatability were docked at a speed of 3,500 per minute. This docking phase, which lasted approximately 90 hours, identified poses with exposed expansion sites for further exploration.
Finally, the top anchor molecules were expanded using in-situ GINGER combined with GPU-accelerated grid/MMFF minimization. This final step was efficiently completed in just 6 hours.
Venetoclax, a groundbreaking BCL-2 inhibitor, stands out with its exceptionally high molecular weight of 868.44 g/mol, contradicting traditional small-molecule drug conventions. Therefore, we thought it would be a challenging example to determine if Venetoclax can be re-built and re-docked using CombiRIDGE. A set of 1M anchors from REAL and a hypothetical Venetoclax synthon were docked and scored. A high scoring pose for the ventoclax anchor was found, and after CombiRIDGE enumeration using GINGER and minimization the re-docking of Venetoclax achieved an impressive RMSD of 1.07 to the crystal structure pose (see below).