Protein Modeling |
Virtual Screening: Ligand and Structure-based |
Lead Optimization |
Cheminformatics Solutions |
Software Development.
The main aim of Molsoft's Contract Research and Consulting Services is to help
in the advancement of our client's research and development objectives through
lasting and collaborative relationships. We provide solutions to specific
scientific problems in the area of drug design, improvements in workflow, and
the management of data, information, and knowledge, tailored directly to our
customers' needs. All our services are undertaken in house by MolSoft's expert research scientists. We use MolSoft's well respected, versatile, and extensive suite of ICM software. Collaborations can be short term, multi-year and are either fee for service or FTE-based. See our
Success Stories site to see some of our published achievements using MolSoft's proprietary ICM software.
Selected Past and Present Collaborations

Accelerating Drug Discovery: Eiletoclax, Lomonotinib, and Balamenib novel new leads in less than 12 months.
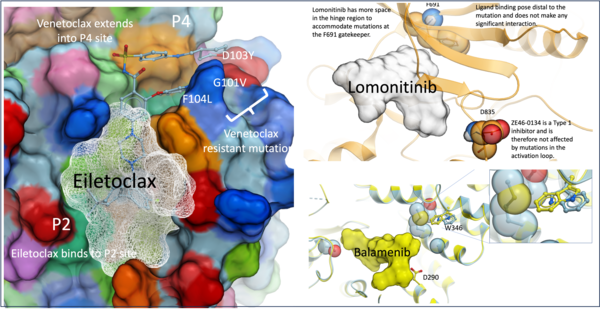
MolSoft is an integral part of the Loch Company Group, maintaining a long-term commitment to providing cutting-edge Computer-Aided Drug Design (CADD) services. The company's strategic focus is centered on oncology and immunology targets, aiming to accelerate the discovery of novel therapeutics.
Our expertise spans the creative design of new therapeutic modalities, including small molecules and hybrid activity compounds. We specialize in addressing challenging drug targets, such as protein-protein interaction (PPI) modulators and PROTACs. In less than a year, our projects have successfully yielded IP-free chemical leads with remarkable potency three projects achieving Ki values below 1nM and four projects below 6nM demonstrating our efficiency in drug discovery.
Several of these compounds, including eiletoclax, lomonotinib, and balamenib, have already entered clinical trials, marking a significant milestone in their development as potential new therapies (see Eilean Therapeutics and Lomond Therapeutics).
Drug Discovery Services
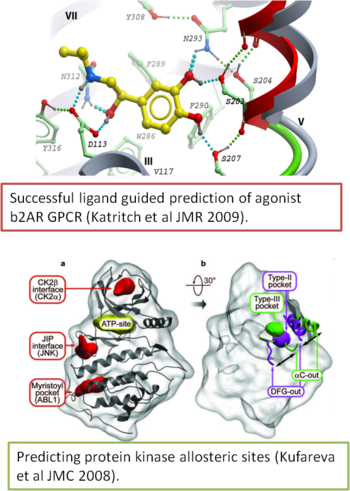
Protein Modeling and Target Prioritization
By applying MolSoft's acclaimed ICM modeling tools we can build three-dimensional models and fully evaluate several candidate target genes.
|
|
 |
Example projects might include:
- Building and optimization of three-dimensional model for each target of interest based on existing homologues with known structure.
- Identifying all potential "drugable" sites and pockets in a protein crystal structure or model.
- Evaluating model errors and structural flexibility around sites of interest.
- Docking all known ligands to the binding sites of a target to validate a ligand binding pocket model.
- Re-docking the ligands to flexible representations of binding sites to improve the pocket models for further virtual ligand screening.
- Investigating the structural effects of mutations on a protein structure.
- Prediction of protein-protein interfaces and interactions using MolSoft's ICM protein-protein docking software. Read about some success stories here.
|
Virtual Ligand Screening Services
The highly specialized research environments of life science industries require
an experienced understanding of the drug discovery process. Many prescription
drugs are the result of small molecules that were synthesized in large
combinatorial libraries and tested ad hoc against potential targets. This
random approach requires substantial capital investment and yields a minimal
number of potential drug candidates. As an alternative to performing
traditional high throughput screening, Molsoft's
Virtual Ligand Screening
methodology can conduct
in silico high throughput screening of multi-million
vendor, multi-billion
ultra-large libraries or combi-chem compound libraries while experimentally testing only a few of the most promising candidates.
MolSoft's screening services provide a comprehensive suite of computational screening solutions to accelerate hit discovery and lead optimization. These services include:
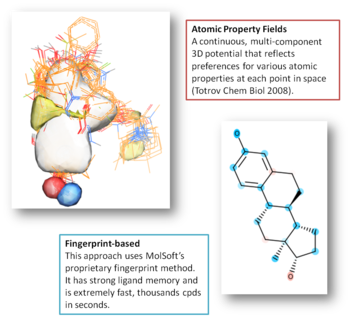
- 3D Ligand-Based Screening - Using Atomic Property Fields (APF) and molecular shape to identify structurally diverse yet functionally similar compounds.
- 3D Structure-Based Docking - High-accuracy flexible docking with MolSoft's ICM docking engine, optimizing ligand poses in target binding sites.
- Synthon-Based Screening - Virtual screening of ultra-large combinatorial libraries (e.g., >28 billion Enamine REAL Space) using the V-SYNTHES method to identify synthetically accessible candidates.
- Custom Virtual Screening Pipelines - Tailored workflows integrating docking, ligand-based methods, and AI-driven optimization.
- Library Preparation & Filtering - Processing, curation, and optimization of commercial and in-house libraries for virtual screening.
- Hit Expansion & Lead Optimization - Computational analoging and scaffold hopping to refine promising hits into optimized lead candidates.
 |
MolSoft's Screening Capabilities
- Molsoft has been a leader in modeling and docking technology and through
continuous development and growth intends to maintain this position.
- MolSoft curates a diverse library of >10 million commercially available compounds and building block fragments for virtual screening. These are off-the-shelf compounds that can be readily purchased for follow-up experimental testing.
- MolSoft's CPU and GPU supercomputing cluster facility enables us to efficiently screen many millions of chemicals per day. Virtual screening on the cloud is also available if extra capacity is needed.
- We can also efficiently undertake screening of Giga Sized libraries using MolSoft's RIDE method, RIDGE, ComnbiRIDGE and Giga Screen methods. Ultra large libraries such as Enamine REAL Space, NIH SAVI, Liverpool Chiro Chem, WuXi Galaxi or Otava CHEMriya can be screened.
- We provide synthon-based screening, enabling virtual exploration of ultra-large combinatorial libraries such as the >28 billion Enamine REAL Space. This approach leverages the ICM-V-SYNTHES method, which efficiently predicts synthetically accessible compounds by assembling virtual molecules from validated building blocks and known reaction rules. This allows for the rapid identification of viable candidates that can be synthesized and tested experimentally.
- MolScreen is a set of high-quality ML/AI 2D fingerprint and 3D pharmacophore models for a wide range of pharmacology and toxicology targets. These models, based on MolSoft's 2D QSAR/Fingerprint and 3D Atomic Property Fields (APF) methods (Totrov, 2008), can be screened as a service for lead discovery or counter-screening, covering approximately 2,500 models across 1,200 targets.
|
 |
Ligand-Based Virtual Ligand Screening
Four types of ligand-based screening are available:
- Fingerprint and substructure-based methods. These methods identify molecular patterns or motifs within compounds, enabling rapid screening by comparing key structural features to known active compounds for better hit prioritization.
- Rapid explicit 3D pharmacophore search. This method searches the spatial arrangement of key pharmacophoric features in a compound's 3D structure, facilitating the quick identification of molecules that match the target's binding profile.
- Shape matching and APF 3D (`apf{Atomic Property Fields}) approach (APF) can be used for chemical superposition, alignment, scaffold hopping and virtual screening.
We have a panel of more than 2500 pre-prepared models of validated drug targets that can be used for lead discovery and prediction of specificity.
|
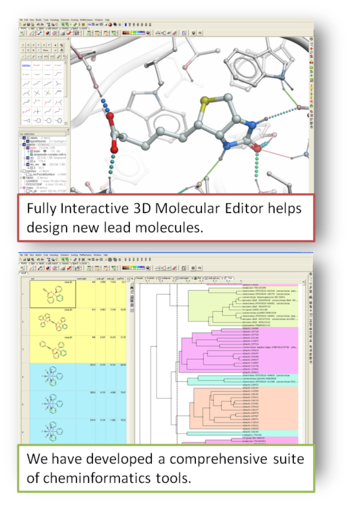
Lead Optimization Services
Once a client has a lead compound, MolSoft provides a number of different services to optimize the drug-like properties of the compound/
|
Projects include:
- Scaffold hopping and lead optimization.
- 3D conformational analysis and generation including peptides and macrocylces.
- QSAR modeling:2D/3D/receptor structure based.
- Cheminformatics: property evaluations, compound clustering, data mining in public and private databases (ChEMBL), library enumeration.
|
Cheminformatics Solutions
Cheminformatics solutions are commonly applied during the lead qualification
and optimization process as it applies to small molecules and proteins.
Molsoft's Cheminformatics applications include structural analysis, simulation
and molecular libraries. Molsoft in a collaboration with
Biovitrum AB, Sweden,
is co-developing a Cheminformatics Client System using Molsoft's proprietary
programming languages and libraries. This product, called BeeHive, is already
being used by Biovitrum and the Structural Genomics Consortium as a program manager and database interface for
chemical searches, analysis, and integration. We also provide services for developing custom database and information management systems. For example, we recently developed the
ChemDiv's Chemistry on Demand E-Shop.
Software Development
Molsoft's Contract Research and Consulting Services division also provides
software development using our libraries of routines for scientific data
analysis and molecular manipulations. Molsoft develops interface software and
the infrastructure necessary to allow communication between instrumentation
controls and the data produced by those controls. The software developed is
able to manage the data generated, to permit access to the databases of
generated data, and to permit visualization and analysis of the data.
Data Communication with the Client

|
Once a milestone has been achieved or the project has been completed, MolSoft's scientists will communicate the results to the cliet in a number of different ways as described below. MolSoft's scientists are available throughout the project to discuss the results and progress either in person, by phone, online via webex.
- Delivery of a single file molecular document with all the results fully annotated and hyperlinked to 3D molecules.
- Delivery of the results in a fully interactive web page using ActiveICM.
- Delivery of screening results in fully-interactive hitlist tables whereby the client can easily view the ligand-receptor complex or lead compound in 2D and 3D.
- Delivery of the results in a portable formate (e.g. a PDB file, SDF file, csv, Excel or high resolution image.
|





