MolSoft ICM RNA Modeling and Lead Discovery |
 A publication in Nature Chemical Biology by scientists at Novartis describes the discovery of a potent small molecule which has the potential to be used to treat Spinal Muscular Atrophy (SMA) a debilitating motor neuron disease. The disease is caused by a deficiency of Survival of Motor Neuron (SMN) gene which is the most common cause of mortality in children. The pyridazine class of compounds described in this paper enhance SMN2 splicing which elevates full-length SMN protein. It is proposed that the small molecule modulators enhance binding of U1 snRNP to the SMN2 exon 7 5'ss.
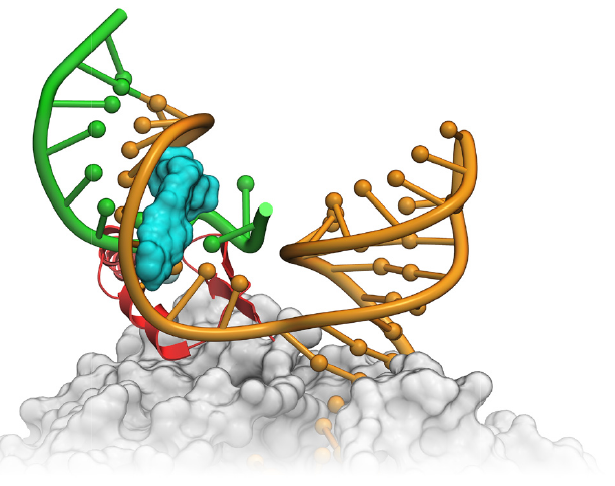
Using MolSoft's ICM Alibero Method, scientists at Novartis Institute of Biomedical Research were able to model the binding mode of the molecule with RNA (See Figure- left). To model the flexibility of RNA a population of 150 RNA models was generated and the Alibero method was able to determine the model that best separated known binders from non-binders. On the basis of this model they were able to hypothesize that the small molecules enhance the stabilization of the RNA duplex in the 1A bulge major groove.
A publication in Nature Chemical Biology by scientists at Novartis describes the discovery of a potent small molecule which has the potential to be used to treat Spinal Muscular Atrophy (SMA) a debilitating motor neuron disease. The disease is caused by a deficiency of Survival of Motor Neuron (SMN) gene which is the most common cause of mortality in children. The pyridazine class of compounds described in this paper enhance SMN2 splicing which elevates full-length SMN protein. It is proposed that the small molecule modulators enhance binding of U1 snRNP to the SMN2 exon 7 5'ss.
Using MolSoft's ICM Alibero Method, scientists at Novartis Institute of Biomedical Research were able to model the binding mode of the molecule with RNA (See Figure- left). To model the flexibility of RNA a population of 150 RNA models was generated and the Alibero method was able to determine the model that best separated known binders from non-binders. On the basis of this model they were able to hypothesize that the small molecules enhance the stabilization of the RNA duplex in the 1A bulge major groove.
Arrakis Therapeutics report a new photoaffinity platform for the analysis of chemical interactions with RNA. The paper in ACS Chemical Biology shows how the method they developed called Photoaffinity Evaluation of RNA Ligation-Sequencing (PEARL-seq) can be used for RNA drug discovery by rapidly identifying ligand binding across many types of RNA. MolSoft's ICM PocketFinder method was used to generate an understanding of where small molecules might bind to Aptamer 21 and then ICM docking was used to predict the docked pose of a probe. The docked pose of the ligand was then used to generate hypotheses around which the binding affinity of the ligand could be improved. The SAR data shows that this method has utility for the design and optimization of new RNA-based small molecule therapeutics.
MolSoft's ICM graphics feature in Chemical and Engineering news article RNA Drug Hunters the https://t.co/t9vlawuQQc Read more about RNA modeling and Lead Discovery using ICM here https://t.co/EhsEsB1EIG #rna #drugdesign #compchem pic.twitter.com/rmWAf0CLz5
— MolSoft LLC (@MolSoft) November 29, 2017
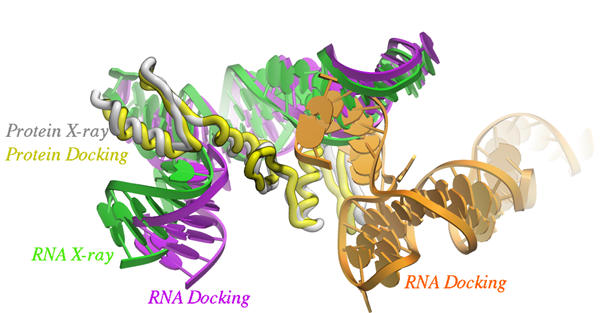 Click here to read paper by Arnautova et al in J Chem Theory Comput. 2018 on MolSoft's ICM Protein-RNA docking method. Paper Abstract: Protein-RNA interactions play an important role in many biological processes. Computational methods such as docking have been developed to complement existing biophysical and structural biology techniques. Computational prediction of protein-RNA complex structures includes two steps: generating candidate structures from the individual protein and RNA parts and scoring the generated poses to pick out the correct one. In this work, we considered three recently developed data sets of protein-RNA complexes to evaluate and improve the performance of the FFT-based rigid-body docking algorithm implemented in the ICM package. An electrostatic term describing interactions between negatively charged phosphate groups and positively charged protein residues was added to the energy function used during the docking step to take into account the greater role that electrostatic interactions play in protein-RNA complexes. Next, the docking results were used to optimize a scoring function including van der Waals, electrostatic, and solvation terms. This optimization yielded a much smaller weight for the solvation term indicating that solvation energy may be less important for the scoring of protein-RNA structures. Rescoring of the generated poses with the new scoring function led to much higher success rates, while pose clustering by contact fingerprints produced further improvements, achieving a success rate of 0.66 for the top 100 structures.
Click here to read paper by Arnautova et al in J Chem Theory Comput. 2018 on MolSoft's ICM Protein-RNA docking method. Paper Abstract: Protein-RNA interactions play an important role in many biological processes. Computational methods such as docking have been developed to complement existing biophysical and structural biology techniques. Computational prediction of protein-RNA complex structures includes two steps: generating candidate structures from the individual protein and RNA parts and scoring the generated poses to pick out the correct one. In this work, we considered three recently developed data sets of protein-RNA complexes to evaluate and improve the performance of the FFT-based rigid-body docking algorithm implemented in the ICM package. An electrostatic term describing interactions between negatively charged phosphate groups and positively charged protein residues was added to the energy function used during the docking step to take into account the greater role that electrostatic interactions play in protein-RNA complexes. Next, the docking results were used to optimize a scoring function including van der Waals, electrostatic, and solvation terms. This optimization yielded a much smaller weight for the solvation term indicating that solvation energy may be less important for the scoring of protein-RNA structures. Rescoring of the generated poses with the new scoring function led to much higher success rates, while pose clustering by contact fingerprints produced further improvements, achieving a success rate of 0.66 for the top 100 structures.