
|
|
2VUW: Eswaran, J., Murray, J.W., Filippakopoulos, P., Soundararajan, M., Pike, A.C.W., Von Delft, F., Picaud, S., Keates, T., King, O., Muller-Knapp, S., Lee, W.H., Marsden, B.D., Wickstroem, M., Bountra, C., Edwards, A.M., Arrowsmith, C.H., Fedorov, O., Burgess-Brown, N., Bray, J., Knapp, S.
3DLZ: Filippakopoulos, P., Eswaran, J., Keates, T., Burgess-Brown, N., Murray, J.W., Von Delft, F., Muller-Knapp, S., Lee, W.H., Marsden, B.D., Arrowsmith, C.H., Edwards, A.M., Wickstroem, M., Bountra, C., Knapp, S.
3F2N: Filippakopoulos, P., Eswaran, J., Keates, T., Burgess-Brown, N., Fedorov, O. Yue, W.W., Murray, J.W., Pike, A.C.W., Von Delft, F., Muller-Knapp, S., Lee, W.H., Marsden, B.D., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Bountra, C. Knapp, S.
3E7V: Filippakopoulos, P., Eswaran, J., Keates, T., Burgess-Brown, N., Fedorov, O., Pike, A.C.W., Von Delft, F., Muller-Knapp, S., Lee, W.H., Marsden, B.D., Arrowsmith, C.H., Edwards, A.M., Wickstroem, M., Bountra, C., Knapp, S.
3FMD: Filippakopoulos, P., Eswaran, J., Keates, T., Burgess-Brown, N., Fedorov, O., Pike, A.C.W., Von Delft, F., Muller-Knapp, S., Lee, W.H., Marsden, B.D., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Bountra, C., Knapp, S.
GSG2/Haspin (haploid germ cell-specific nuclear protein kinase) is a serine/threonine kinase that tightly associates with chromosomes during mitosis. Kinases of this family share only very low sequence homology with other protein kinases, contain several large insertions and lack a subset of residues that are almost invariant in known kinases [1]. Phosphorylation of Haspin itself and its specific substrate histone H3 threonine-3 (H3T3ph) occurs during mitosis. Haspin has been shown to be vital for the maintenance of chromosome cohesion. Depletion of Haspin leads to a loss of cohesin association, premature chromatid separation, activation of the spindle assembly checkpoint, and a block in mitosis in a pro-metaphase-like state [2]. Although no direct link to cancer has been established so far, Haspin has been suggested as a target for drug development as it has been shown to activate the well established oncology target Aurora 2 [3]. Here we present the structure of the Haspin kinase domain in complex with ATP and ATP mimetic inhibitors.
Structural Features
The crystallized constructs comprises Haspin residues Lys470-Lys798. The first crystals were obtained with the adenosine kinase inhibitor 5-Iodotubercidin (4-Amino-5-iodo-7-(b-D-ribofuranosyl) pyrrolo[2,3-d]pyrimidine). This structure was solved by SAD and was refined at 1.8 resolution. The kinase domain contains several inserts not present in other protein kinases which include an alpha helical insert between the beta strands 4 and 5, an unique activation segment and a large beta-hairpin insert in the lower kinase lobe.
(TIP 1: the background colour can be changed to [WHITE] or [BLACK] )
(TIP 2: reset the view )
The alpha helical insert in the upper kinase lobe (residues Phe593-Tyr569) binds to a deep cradle formed by the 5 beta strands. Its conformation is stabilized by a number of hydrophobic and aromatic contacts and this haspin unique motif is likely to stabilize the P-loop in an active conformation.
The catalytically important helix C is embedded in a hydrophobic pocket partially formed by a unique beta hairpin activation segment insert. The conserved salt bridge between the C glutamate (Glu535) and the active site lysine residue (Lys511) typically found in active kinases is formed indicating the correct positioning of C for catalysis.
The haspin activation loop comprises a number of unique features: i. the conserved ATP/Mg binding motif DFG is changed to DYT , ii. there is a beta hairpin insert that interacts and stabilizes C, iii. a cluster of acidic residues (Asp707, Glu708, Asp709) form a negatively charged substrate binding channel, iv. an aromatic core stabilizes the conformation of the entire segment, v. the catalytic loop HRD motif arginine (Arg648), that often interacts with phosphate moieties in kinases that require activation segment phosphorylation, interacts with the backbone of the activation loop tip and vi. the missing APE motif is replaced by a larger helical insert (Gln718-Lys727).
A haspin specific hairpin insert (Lys663-Cys679) packs against the lower lobe and makes a number of main chain contacts with a loop region C-terminal to C. A similar long hairpin has also been observed in CLK kinases but in CLK1 and CLK3 this secondary structure element is oriented differently and packs against opposite sides in the lower kinase lobe.
The electrostatic potential of the haspin surface suggests how the basic histone tail of H3 binds to the substrate binding site. It is quite likely that the strongly negatively charged channel formed by the activation segment accommodates the histone tails while the basic histone docks at the upper lobe of the kinase.
Haspin has a very unique ATP binding site and some motifs known to be important for the binding of Mg/ATP (e.g. the DFG) are altered. To understand how ATP binds to this changed active site we crystallized Haspin with ATP/Mg. However, the ATP hydrolyzed after the alpha-phosphate and only AMP was visible in the electron density map . The Adenosine ring forms the typical hinge interactions with the hinge backbone residues (Gly609 and Glu606). Interestingly, we detected a so far unseen Mg-binding site in the interface between the P-1-loop, the catalytic loop and F .
The unique active site properties may also lead to the development of specific inhibitors. We crystallized Haspin with 5-Iodotubercidin (4-Amino-5-iodo-7-(b-D-ribofuranosyl)), and ATP mimetic inhibitor that has originally been described as an Adenosine kinase inhibitor. The bulky iodide moiety forms a tight interaction with the aromatic ring of the Phe605 gatekeeper. In addition, 5-Iodotubercidin forms two hydrogen bonds with the hinge (Gly608 and Glu606).
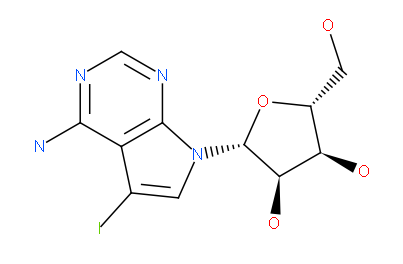
5-Iodotubercidin (4-amino-5-iodo-7-(D-ribofuranosyl) pyrrolo[2,3]pyrimidine)
We also solved the structure of haspin in complex with an imidazo[1.2-b]pyridazine and an pyrazolo[1.5a]pyrimidine inhibitor. Both inhibitors have very good shape complementarity and similar binding modes and form an aromatic -stacking interaction with the Phe605 gatekeeper and a polar contact with the hinge backbone (Glu606).
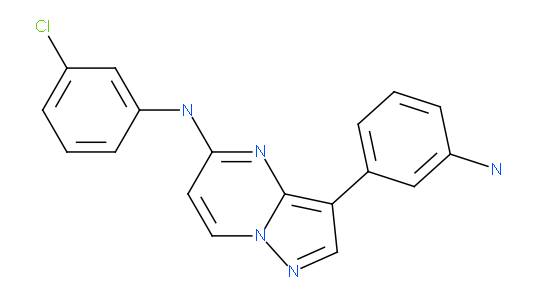
pyrazolo[1.5-a]pyrimidine
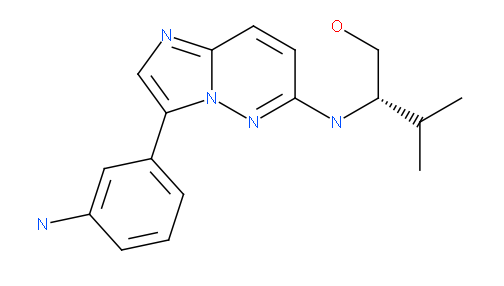
imidazo[1.2-b]pyridazine
We also solved the structure of haspin in complex with the ATP mimetic isoquinoline inhibitor . This inhibitor scaffold was originally designed to inhibit PKC kinases and is quite selective for PKCs and gsg2. Despite the potency of the inhibitor (IC 50 : 160 nM) the inhibitor is not well optimize for the haspin binding pocket and there are multiple possibilities for improvement.
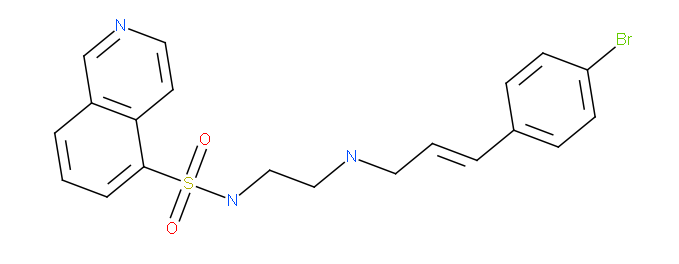
isoquinoline
Note: The target annotations and structure descriptions within this datapack are compiled by our Principal Investigators and are not peer-reviewed. If you find anything in the annotations that is not accurate, please notify us using the our on-line feedback page or send an e-mail to contact us .
1. Higgins, J.M. (2001). Haspin-like proteins: a new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Science 10, 1677-1684.
2. Dai J., Sultan S., Taylor S.S., Higgins J.M. (2005). The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes and Development 19 , 472-488.
3. Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. (2008). Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science 319 , 469-472.
Datapack created using Molsoft ICM and Molsoft Browser technologies (patent pending).