|
Substrate Specificity In Human Monomeric Carbonyl Reductases - Pilka et al .
Table 1: Activity of CBR1 and CBR3 against selected substrates. The threshold for significant activities (measured at 200 μM substrates and 200 nM (~ 6.6 μg/ml) enzyme concentration) was set to 10% of the activity of CBR3 against 1,2-naphthoquinone (2.5 μmol/(min mg)), which was set to 1.0 for comparison. All other activities denoted either b.t. (below threshold) or n.a. (no activity observed).
|
|
Structure |
Relative CBR1 activity |
Relative CBR3 activity |
|
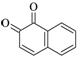
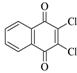
1,2-Naphthoquinone |
|
1.70 |
1.00 |
|
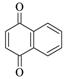
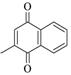
1,4-Naphthoquinone |
|
2.5 |
b.t. |
|
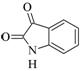
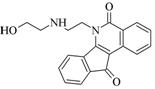
Isatin |
|
0.39 |
0.15 |
|
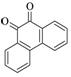
9,10-Phenanthrenequinone |
|
7.90 |
0.19 |
|
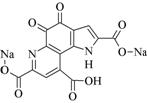
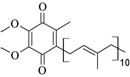
Methoxatin; Pyrroloquinoline quinone |
|
1.69 |
0.30 |
|
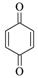
1,4-Benzoquinone |
|
0.30 |
b.t. |
|
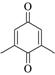
2,6-Dimethyl-p-benzoquinone |
|
0.80 |
b.t. |
|
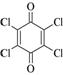
Chloranil |
|
1.49 |
b.t. |
|
Coenzyme Q10 |
|
0.35 |
b.t. |
|
Menadione |
|
1.20 |
b.t. |
|
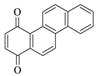
1,4-Chrysenequinone |
|
0.5 |
b.t. |
|
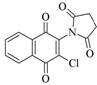
2-Chloro-3-(N-succinimidyl)-1,4-naphthoquinone |
|
2.92 |
0.30 |
|
Dichlon |
|
0.89 |
n.a. |
|
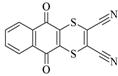
Dithianon |
|
0.67 |
b.t. |
|
6-Anilinoquinoline-5,8-quinone |
|
1.43 |
b.t |
|
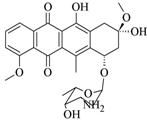
Daunorubicin (Daunomycin) |
|
0.26 |
b.t. |
|
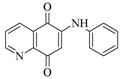
Oracin |
|
n.d.* |
0.14* |
|
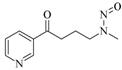
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) |
|
0.09 |
b.t. |
|
Erbstatin analog |
|
0.12 |
0.13 |
|
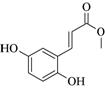
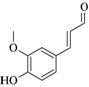
Coniferyl aldehyde |
|
0.30 |
0.18 |
|
Acetohexamide |
|
0.11 |
0.19 |
* activity of CBR3 towards oracin measured by HPLC, no measurement for CBR1
Table 2: Comparison of kinetic constants for human CBR1 and CBR3 against selected substrates.
|
Substrate |
CBR1 activity |
CBR3 activity |
||
|
|
Km [μM] |
Vmax
|
Km [μM] |
Vmax
|
|
1,2-Naphthoquinone |
310 |
11 |
420 |
6 |
|
1,4-Naphthoquinone |
560 |
10 |
n.a.* |
n.a.* |
|
9,10-Phenanthrenequinone |
35 |
9 |
> 80** |
< 0.1** |
|
Isatin |
2 |
2 |
14630 |
15 |
|
Oracin |
n.d.*** |
n.d.*** |
140 |
0.1 |
|
NNK |
7500 |
3 |
n.a.* |
n.a.* |
* very little activity detected
** no Michaelis-Menten kinetic observed, value estimated from the slope of linear regression of the relation between activity and substrate concentration
*** no literature data
Table 3: Relative catalytic efficiency (kcat/ K m) of CBR1/ CBR3 mutants and wild-type proteins against naphthoquinone substrates. Catalytic efficiency of WT CBR3 against 1,2-naphthoquinone was set to 1.0 for comparison.
|
CBR1 |
1,2-Naphthoqui-none [ortho-] |
1,4-Naphthoqui-none [para-] |
CBR3 |
1,2-Naphthoqui-none [ortho-] |
1,4-Naphthoqui-none [para-] |
|
WT |
13.3 |
6.4 |
WT |
1.0 |
< 0.1 |
|
W229F |
10.4 |
2.1 |
P230F |
0.3 |
0.3 |
|
W229P |
8.8 |
0.5 |
P230W |
0.2 |
0.2 |
|
W229P/A235D |
1.5 |
< 0.1 |
P230W/D236A |
0.7 |
0.1 |
|
W229P/M141A |
n.d.* |
n.d.* |
P230W/Q142A |
0.4 |
< 0.1 |
|
W229P/M141Q |
n.d.* |
n.d.* |
P230W/Q142M |
0.7 |
< 0.1 |
|
M141Q |
9.7 |
3.5 |
Q142M |
1.2 |
0.3 |
|
M141A |
8.5 |
1.8 |
D236A |
< 0.1 |
0.1 |
* protein unstable
Table 4: Comparison of kinetic constants for selected CBR1/ CBR3 mutants and wild-type proteins against isatin.
|
CBR1 |
Km
|
Vmax
|
CBR3 |
Km
|
Vmax
|
|
WT |
1.66 ± 0.45 |
2.05 ± 0.04 |
WT |
14630 ± 54 |
14.75 ± 0.06 |
|
W229F |
5.13 ± 2.1 |
10.46 ± 0.7 |
P230F |
15419 ± 4798 |
7.78 ± 2.0 |
|
W229P |
40.22 ± 4.9 |
19.55 ± 0.76 |
P230W |
n.t.* |
n.t.* |
|
W229P/A235D |
2930 ± 248 |
116.90 ± 5.2 |
P230W/D236A |
5084 ± 420 |
19.90 ± 1.05 |
|
|
|
|
D236A |
3570 ± 599 |
37.68 ± 3.6 |
* not tested
Download Standalone iSee datapack: You can download and view all the Information of a datapack offline including information not available in the web version (where applicable). You will also need to download and install the ICM-Browser to view the standalone datapacks.


Datapack created using Molsoft ICM and Molsoft Browser technologies. (Patent Pending)