The Latest Molsoft News |
Click here to subscribe to the MolSoft News Mailing List (approx 1 Email/month) or follow us on: Twitter, Facebook or YouTube.
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001 | 2000 | 1999 | Founded in 1994 |

San Diego, CA — November 12th 2025 MolSoft LLC announces GigaSearch v2.0, a next-generation chemical search engine capable of scanning billions to trillions of compounds within seconds. Built for ultra-large substructure, similarity and analog searches, GigaSearch v2.0 enables rapid exploration of vast chemical spaces such as Enamine REAL, Chemspace Freedom, and other make-on-demand libraries.
GigaSearch v2.0 can be accessed directly via the MolSoft server through the Chemical Search → Online Databases tab in ICM-Pro and ICM-Chemist, or installed locally under license for in-house high-speed screening.
Read more here.
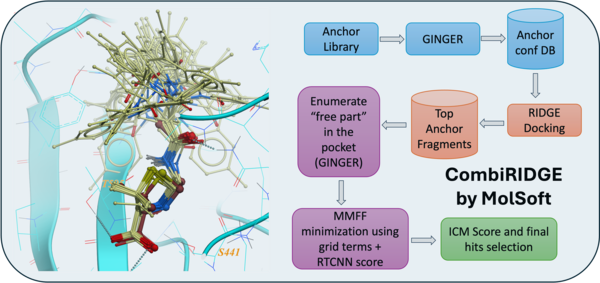
San Diego, CA — October 1st 2025 — MolSoft LLC today announced the launch of CombiRIDGE, a GPU-accelerated combinatorial docking and virtual screening solution designed to efficiently explore ultra-large chemical spaces. CombiRIDGE combines neural networks, fast GPU docking, and modern scoring functions to accelerate hit discovery and lead optimization.
CombiRIDGE enables drug hunters to optimize and evaluate R-groups around a defined core fragment, referred to as an anchor, to rapidly generate and score complete molecules directly in a protein binding site. Rather than enumerating full libraries in advance, CombiRIDGE grows combinatorial libraries on-the-fly, allowing efficient navigation of extremely large chemical spaces.
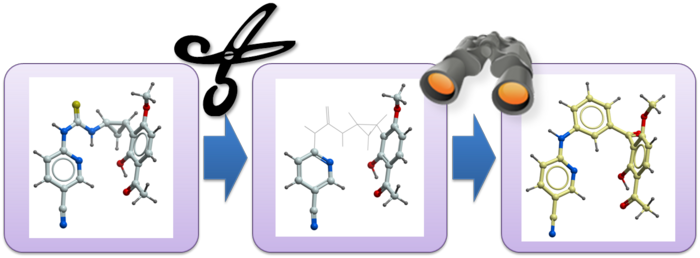
GINGER is MolSoft’s neural network-based conformer generation technology. It generates accurate three-dimensional conformations of anchor fragments directly in the context of the protein binding site, ensuring that downstream docking starts from realistic ligand geometries.
RIDGE is MolSoft’s GPU-optimized docking engine. It enables extremely fast docking of millions of anchor conformations, making it possible to screen very large libraries using a small number of modern GPUs.
RTCNN is a graph neural network-based scoring function trained to evaluate protein-ligand interactions. RTCNN provides robust ranking of docking poses and helps prioritize high-quality candidates for further expansion and experimental follow-up.

Molsoft scientists have successfully developed and deployed powerful GPU-accelerated virtual screening engines to rapidly screen chemical libraries containing billions of compounds. In collaboration with Nadya Tarasova's group at the NIH this technology has led to the discovery of potent drug leads against two historically "undruggable" cancer targets, K-Ras G12D and PD-L1. Read the paper here.
The ability to screen massive chemical spaces is vital for next-generation drug discovery. Molsoft's GPU implementations achieve this with unprecedented speed:
The publication reports novel GPU-accelerated screening methods that significantly outperform conventional virtual ligand screening (VLS). MolSoft's docking scores proved to be a more accurate predictor of binding, leading to the successful identification of eight potent inhibitors (five for PD-L1, three for K-Ras G12D) with submicromolar affinities. These powerful new methods effectively expand the range of molecules MolSoft can screen and enable hit identification even for challenging drug targets.
MolSoft today released a white paper about the RTCNN - Radial and Topological Convolutional Neural Network Score which is outperforming other Virtual Screening scoring methods. The RTCNN score is used in all MolSoft's structure-based screening methods including ICM-VLS, RIDGE, GigaScreen and CombiRIDGE.
Ligand scoring plays a crucial role in virtual screening, a computational technique used to identify potential drug candidates from large compound libraries. Graph Convolutional Neural Networks (GCNNs) have shown promise in improving ligand scoring by capturing the structural and chemical features of molecules in a graph representation. Traditional ligand scoring methods rely on handcrafted features and simplified physical energy terms that may not fully capture the complex interactions between a ligand and its target protein. GCNNs offer a data-driven approach by learning directly from the molecular graph representation, allowing for more accurate and robust scoring.
Read More...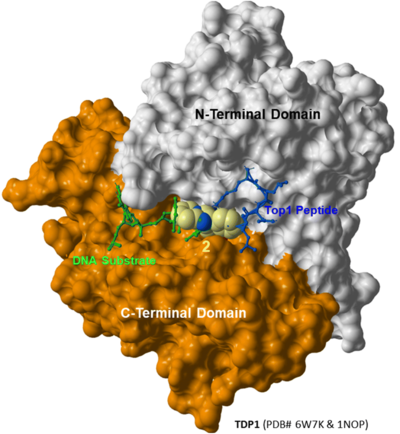 A new study published today in RSC Medicinal Chemistry by Dr. Burke's lab at the NIH National Cancer Institute (NCI) explores the development of proteolysis-targeting chimeras (PROTACs) to selectively degrade tyrosyl-DNA phosphodiesterase 1 (TDP1). The paper, titled "Application of a Bivalent "Click" Approach to Target Tyrosyl-DNA Phosphodiesterase 1 (TDP1)," highlights the potential for PROTACs to enhance the efficacy of topoisomerase 1 (TOP1) inhibitors used in cancer therapy.
A new study published today in RSC Medicinal Chemistry by Dr. Burke's lab at the NIH National Cancer Institute (NCI) explores the development of proteolysis-targeting chimeras (PROTACs) to selectively degrade tyrosyl-DNA phosphodiesterase 1 (TDP1). The paper, titled "Application of a Bivalent "Click" Approach to Target Tyrosyl-DNA Phosphodiesterase 1 (TDP1)," highlights the potential for PROTACs to enhance the efficacy of topoisomerase 1 (TOP1) inhibitors used in cancer therapy.
Using MolSoft ICM, the researchers modeled the ternary structure of a PROTAC complex, docking a lead imidazopyridine-based TDP1 inhibitor (compound 1b) with thalidomide to form a bivalent conjugate (compound 5h). The modeling results confirmed that the linker in 5h was optimally positioned, extending from the peptide-binding groove of TDP1 toward the CRBN binding site, supporting the intended PROTAC mechanism.
This study provides valuable insights into the structure-based design of TDP1-targeting PROTACs, paving the way for future therapeutic advancements.
 Advent Informatics, the distributor of MolSoft's ICM-Pro software in India, recently hosted a well-attended and engaging User Group Meeting. The event featured compelling success stories in areas such as small-molecule drug design and discovery, PROTAC development, and antibody design. It also showcased innovative approaches for ultra-large library screening, multiple receptor docking, and covalent docking. The meeting included insightful presentations by scientists from leading pharmaceutical and biotech companies. The following speakers presented their research:
Advent Informatics, the distributor of MolSoft's ICM-Pro software in India, recently hosted a well-attended and engaging User Group Meeting. The event featured compelling success stories in areas such as small-molecule drug design and discovery, PROTAC development, and antibody design. It also showcased innovative approaches for ultra-large library screening, multiple receptor docking, and covalent docking. The meeting included insightful presentations by scientists from leading pharmaceutical and biotech companies. The following speakers presented their research:
|
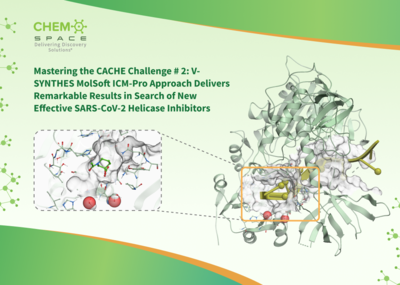
MolSoft LLC is pleased to share that ChemSpace has completed the CACHE 2 Challenge with notable success, utilizing MolSoft's ICM software suite. Leveraging MolSoft's ICM tools, Chemspace achieved impressive results in high-throughput virtual screening, significantly advancing the discovery of promising drug candidates for the COVID target NSP13 helicase.
The results highlights the capabilities of MolSoft's ICM platform, which enabled Chemspace to screen large ligand libraries (176B chemicals) accurately and at high speed. This outcome demonstrates the robustness of MolSoft's technology in screening ultra-large libraries, enabling efficient and accurate identification of high-quality hits Read more on ChemSpace's website here. |
|
A paper by MolSoft's scientists and developers Eugene Raush (Princpal Developer), Ruben Abagyan (MolSoft Founder) and Maxim Totrov (CTO) has today been published in the ACS Journal of Chemical Theory and Computation. The paper unveils a novel cutting-edge algorithm called GINGER, which efficiently generates high quality chemical conformers at lightning-fast speeds. This innovative approach leverages neural networks to rapidly predict low-energy conformers from 2D chemical structures. GINGER's efficiency is further enhanced by GPU optimization, enabling rapid processing of large compound libraries.
GINGER can convert 150-200 mol/sec on a single Nvidia RTX 4090 GPU making it suitable for the efficient conversion of ultra-large libraries (>1B molecules) when run on a multi-GPU server. The ultra-large libraries can be screened using MolSoft's rapid field-based 3D similarity search engine RIDE or via fast structure-based docking use RIDGE and GigaScreen.
GINGER is a separate add-on module to MolSoft's desktop modeling software ICM-Pro and ICM-Chemist-Pro or can be run by MolSoft as a service. Please contact MolSoft with any questions and to obtain an evaluation license or to purchase GINGER.
|

|
MolSoft and Liverpool Chiro Chem (LCC) have partnered to make the LCC's 1.4 Billion ultra-large chemical library available for virtual screening. The database can be screened rapidly via structure or ligand based approaches using MolSoft's unique RIDGE and RIDE flexible docking algorithms ported to modern GPU processors.
LCC's ultra-large library has been converted to 3D conformers using Molsoft's new highly accurate neural network GINGER conformer generator. The database is based on LCC's proprietary collection of chirally pure, sp3-rich scaffolds with a diverse set of ring sizes, ring topologies, and exit vectors. The reaction-based enumeration facilitates transformation of your chiral virtual hits to high-quality physical samples.
MolSoft offers a cost-effective service for rapidly screening the database, with results analyzed by our team of computational chemistry experts. Virtual hits then can be converted to physical compounds using LCC's efficient parallel synthesis laboratories. The library can be screened by MolSoft either through a structure- or ligand-based approach as a service.
Structure-Based Lead Discovery
If atomic resolution structures or AlphaFold models of your target protein or nucleic acid are available, screening the LCC database against the structure can be undertaken using MolSoft's high-performance GPU-based RIDGE-docking or deep learning GigaScreen methods. The top scoring compounds will undergo inspection, and a ranked hitlist will be provided along with 3D poses of the top hits in complex with the receptor along with other essential properties.
Ligand-Based Isostere Scaffold Hopping Discovery If one or more lead chemicals are available, the library can undergo screening using MolSoft's Rapid Isostere Discovery Engine (RIDE) approach. This will enable new chemistry with similar 3D pharmacophore properties and shape to be identified from the LCC database. Atom weighting can be employed to highlight the significance of specific moieties, while excluded volumes penalties can be applied to regions surrounding the query molecule, prioritizing hits without bulky extensions in constrained areas.
MolSoft's screening methods are finely tuned for high-performance GPUs, allowing us to swiftly deliver screening results to your company within 1-2 weeks in most cases.
Please contact MolSoft if you have any questions or wish to schedule a call to discuss your project and MoSoft's virtual screening approach of the LCC database. Please contact LCC if you have any questions regarding the ultra-large chemical library.
About Molsoft Molsoft LLC is a leading provider of Physics- and AI- based scientific software solutions, specializing in computational chemistry, biology, and molecular modeling. MolSoft's flagship product, ICM (Internal Coordinate Mechanics), equips researchers in academia, pharmaceuticals, and biotechnology with powerful tools for molecular design, virtual screening, and drug discovery. With a dedication to in silico method innovation, Molsoft is committed to advancing the frontiers of scientific research and pharmaceutical development.
About Liverpool ChiroChem LCC is an established, chemical technology innovator, on a mission to accelerate the discovery and development of high-quality drugs. LCC provide expanded access to 3D chemical space to support orthogonal hit-ID (DEL, FBLD, VLS), hit optimisation using our parallel synthesis laboratory, and personalised support using our design and computational team to expedite targeted chemical space exploration.

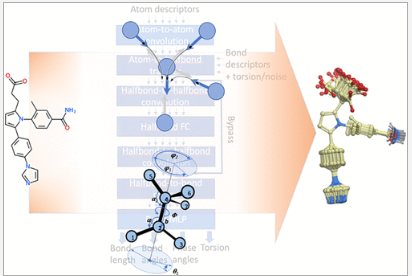
Conformer generation is an essential step of a variety of molecular modeling and computer-assisted drug discovery workflows such as 3D ligand-based virtual screening or fast GPU docking. GINGER (Graph Internal-coordinate Neural-network conformer Generator with Energy Refinement) is Molsoft's new cutting-edge software designed for lightning-fast high quality conformer library generation on GPUs.
GINGER is available as an add-on to ICM-Pro and ICM-Chemist-Pro. Please contact us if you have any questions or if you would like to add GINGER to your current ICM configuration.
Read more....

 Eilean Therapeutics LLC is a company that utilizes MolSoft's AI and Physics-Based molecular modeling tools to rationally develop first and best in class therapeutics.
Eilean Therapeutics LLC is a company that utilizes MolSoft's AI and Physics-Based molecular modeling tools to rationally develop first and best in class therapeutics.
You can read Eilean Therapeutic's recent press release below: Eilean Therapeutics LLC, a biopharmaceutical company dedicated to discovering and developing best-in-class and first-in-class small molecule inhibitors to target escape mutations in hematologic malignancies, today announced the initiation of human dosing in the Phase 1 clinical program of ZE46-0134, a highly potent and selective pan-FLT3 inhibitor that targets clinically relevant FLT3 mutations including the FLT3 gatekeeper mutation, while sparing the wild type form of the protein. Read More...
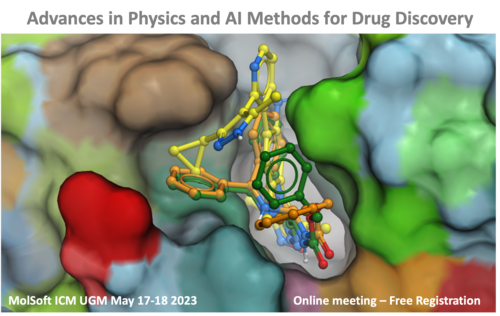
On May 17-18, 2023 MolSoft hosted its highly anticipated User Group Meeting in an online format. The event featured engaging talks by both our esteemed users and talented developers, spotlighting the exciting array of features present in ICM. Attendees learnt about the latest cutting-edge technologies, including GPU-based and AI-accelerated virtual screening, structure- and ligand-based drug design, PROTAC modeling, membrane protein modeling, and much more. You can watch a selection of the presentations again on our YouTube channel - see the links below:

Prof. Ruben Abagyan (UCSD, MolSoft Founder) presented at and chaired the panel for the session "Artificial Intelligence for Early Drug Discovery" at the Drug Discovery Chemistry Conference in San Diego this week. The title of his presentation was "Giga-Screening for Preclinical Candidates with a Defined Multi-Target Profile" and his talk discussed the challenges involved with drug discovery of "tough targets" such as FLT3 and BCL2. Rational design of first- and best- in class candidates was achieved under two years using MolSoft's AI and Physics Based molecular modeling tools. The candidates for primary and drug resistant leukemias and lymphomas are initiating clinical studies. You can read more about this work at i2020 Accelerator here.
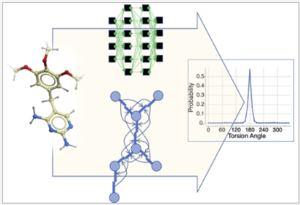 A new MolSoft ICM paper has been published in the J.Chem.Inf.Model - you can read it here. The paper describes a new graph-convolutional neural network (GCNN)-based method which can be used for learning and prediction of statistical torsional profiles (STP). The model has been trained on the Crystallography Open Database and has good correlation with quantum chemically calculated profiles. The method can be used for conformer generation and you can try the method here.
A new MolSoft ICM paper has been published in the J.Chem.Inf.Model - you can read it here. The paper describes a new graph-convolutional neural network (GCNN)-based method which can be used for learning and prediction of statistical torsional profiles (STP). The model has been trained on the Crystallography Open Database and has good correlation with quantum chemically calculated profiles. The method can be used for conformer generation and you can try the method here.

At MolSoft we have been privileged to collaborate with many exceptionally talented Ukrainian scientists and we aim to preserve those ties during this tragic time in Ukraine. Ukrainian companies such as Enamine and ChemSpace are currently relocating to other countries so as to keep serving their customers. We believe that one way the MolSoft ICM community can support them during this time is by maintaining the use of their products and services as normal, and providing them with our services.
You can now use MolSoft's Rapid Isostere Discovery Engine ( RIDE) method to efficiently screen over 48 billion chemical conformers from the Enamine REAL database. This will allow you to find new leads and scaffold hop to new interesting chemistry to aid your drug discovery program and quickly find new drugs for many kinds of diseases. MolSoft has collaborated with the NIH and Enamine to prepare Enamine's REAL database for virtual screening . The REAL database is a reaction enumerated library containing approximately 2 billion chemicals (read more here).
Ruben Abagyan Ph.D. (MolSoft Founder) said "We are excited to collaborate with Enamine and the NIH on another Giga sized screening project. RIDE is already being used routinely by our customers to screen the Giga-sized NIH SAVI library and we have worked with Enamine in the past to develop an efficient search method for REAL. We are happy that our customers will have another option for their drug discovery needs."
Nadya Tarasova Ph.D. (NIH/NCI) said "It was a big scientific and computation challenge to prepare the REAL library conformer database and it took more than 7 million CPU hours to complete but now the library is ready we are excited to see how it will be used by the scientific community to deliver new drugs to the market. We have already started screening the REAL database using MolSoft's RIDE method for drug discovery projects here at the NIH."
Vladimir Ivanov Ph.D. (Chief Sales and Marketing Officer at Enamine) said "We are happy that MolSoft's ICM user community will have access to efficiently screen the REAL database using the RIDE method. At Enamine we have enabled a high synthesis success rate for the REAL molecules and can make them available within 3-4 weeks. We look forward to working with MolSoft and the ICM users to help provide chemicals for their drug discovery projects."
MolSoft's RIDE is a fast 3D molecular similarity search method based on Atomic Property Fields (APF) invented by Molsoft Lead Scientist, Max Totrov, and implemented by Molsoft Lead Software Engineer Eugene Raush and his team. RIDE searches databases of compound conformers for molecules that are isosteric to the query, i.e. have similar 3D configurations and distributions of atomic properties. RIDE is a multi-conformational optimal 3D superposition in which both the query molecule and the molecule in the database are represented in multiple low energy conformations to reflect the flexible nature of both molecules. RIDE is available in the ICM-Pro + VLS package.
Applications screening the REAL database with RIDE include:
If you want to learn more about RIDE please see a recorded webinar here on YouTube.
The REAL database is saved in MolSoft's compact MOLT file format - please contact MolSoft for download instructions. Other RIDE formatted databases, including the NIH Giga Sized SAVI database are available here.
 At MolSoft we have been privileged to collaborate with many exceptionally talented Ukrainian scientists and we aim to preserve those ties during this tragic time in Ukraine. Ukrainian companies such as Enamine and ChemSpace are currently relocating to other countries so as to keep serving their customers. We believe that one way the MolSoft ICM community can support them during this time is by maintaining the use of their products and services as normal, and providing them with our services.
At MolSoft we have been privileged to collaborate with many exceptionally talented Ukrainian scientists and we aim to preserve those ties during this tragic time in Ukraine. Ukrainian companies such as Enamine and ChemSpace are currently relocating to other countries so as to keep serving their customers. We believe that one way the MolSoft ICM community can support them during this time is by maintaining the use of their products and services as normal, and providing them with our services.
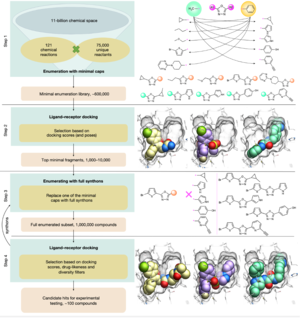 MolSoft's ICM accurate flexible docking, cheminformatics and virtual screening technology provided the unique and powerful engine for a successful screen of as many as 11 billion chemical compounds, potential drug candidates. The USC laboratory of Prof. Vsevolod Katritch report in Nature an approach called V-SYNTHES. The tool performs a ICM-docking based hierarchical structure-based screening of a Enamine REAL Space giga library. The screen resulted in the discovery of new lead compounds for the GPCR CB2 and the Kinase ROCK1.
MolSoft's ICM accurate flexible docking, cheminformatics and virtual screening technology provided the unique and powerful engine for a successful screen of as many as 11 billion chemical compounds, potential drug candidates. The USC laboratory of Prof. Vsevolod Katritch report in Nature an approach called V-SYNTHES. The tool performs a ICM-docking based hierarchical structure-based screening of a Enamine REAL Space giga library. The screen resulted in the discovery of new lead compounds for the GPCR CB2 and the Kinase ROCK1.
For many years, MolSoft has been developing methods for fast accurate docking and scoring tools, and incorporating those tools to search giga-sized libraries. These tools include, ICM-Pro docking, GIGA search, and a shape property-based pharmacophoric search method called RIDE. GIGA search and RIDE were applied to the NIH SAVI database and Enamine REAL database.
For V-SYNTHES, ICM-Pro was used as the docking engine as well as for the preparation of the reaction libraries and enumeration of the giga-sized virtual databases. Docking and screening simulations were then undertaken using MolSoft's ICM-Pro Virtual Ligand Screening tools. The docking uses MolSoft's Biased Probability Monte Carlo method, optimized force field, docking score to dock and score fragments of a combinatorial library, and then an empirical score is derived and applied to a much larger library. At the end, the top scoring hits from the library are re-docked and scored again. This hierarchical protocol accelerates the docking and scoring run and makes it applicable to very large databases. The flexibility of the binding pocket was represented using MolSoft's 4D docking methodology which uses an ensemble of structures efficiently for fast and accurate flexible-receptor ligand docking.
Prof. Katritch at USC says "We are very excited to use the MolSoft ICM-Pro technology to address the challenges of screening very large chemical databases for the discovery of drug candidates."
You can view and download additional ICM scripts used for this work at the Katritch lab GitHub site which are ready for use with the ICM-Pro + VLS desktop modeling software.
Please contact MolSoft (www.molsoft.com info@molsoft.com) if you have any questions about how to apply this approach to your drug discovery program.
Announcing new start-up investments aimed to bring proprietary small molecule products to the clinic targeting cancer together with @OrbiMed! https://t.co/klV76hoZee #lifescienceindustry #startupecosystem #biotechnology #oncologyclinicalresearch #cancertreatment #accelerator pic.twitter.com/d1QjdL6ljQ
— i2020accelerator (@i2020accelerat1) September 14, 2021

MolSoft has collaborated with the NIH/NCI (Tarasova and Nicklaus labs at NIH/NCI) to make their Synthetically Accessible Virtual Inventory (SAVI) database available to screen using MolSoft's Rapid Isostere Discovery Engine ( RIDE). You can read more about how the NIH developed SAVI in this publication here. The SAVI database contains more than 1 billion chemicals and RIDE can screen 500K chemical conformations/sec/GPU so the database can be screened very quickly.
RIDE is a fast 3D molecular similarity search method based on Atomic Property Fields (APF). RIDE searches databases of compound conformers for molecules that are isosteric to the query, i.e. have similar 3D configurations and distributions of atomic properties. RIDE is a multi-conformational optimal 3D superposition in which both the query molecule and the molecule in the database are represented in multiple low energy conformations to reflect the flexible nature of both molecules. RIDE is available in the ICM-Pro + VLS package.RIDE applications include:
If you want to learn more about RIDE please see a webinar here https://youtu.be/oA6T3W22r5c
You can download the SAVI database and other databases formatted for RIDE here http://www.molsoft.com/addons/conf/ Please be aware that the SAVI database is 4.3 Terabytes in size so will take a while to download and we recommend for optimal performance the database should be located on local SSD or fast HDD.
Happy to announce additional financing for our breakthrough Cancer-, Inflammation- and Neurology-focused therapeutics together with @OrbiMed! Learn morehttps://t.co/0z7McltyGv #startup #accelerator #LifeSciences #Cancer #Investment #neurology pic.twitter.com/hX5ZnlmWzp
— i2020accelerator (@i2020accelerat1) April 29, 2021
Mcule's ULTIMATE library is now available in Molsoft's ICM-Chemist and ICM-Pro desktop modeling packages. ULTIMATE contains more than 100 million drug-like chemicals which can now be searched using 2D and 3D pharmacophore methods developed by MolSoft. As more than 95% of the compounds in ULTIMATE are completely novel, researchers can easily find new chemical starting points and move away from patent space.
ICM allows users to search ULTIMATE using MolSoft's MolCart Giga-Search technology. It can be searched using chemical substructure, patterns/fingerprints, similarity, exact match, and the results can be analyzed and filtered using Multi Parameter Optimization (MPO). Structure-based and fast ligand-based virtual screening of ULTIMATE can also be undertaken in the ICM-Pro software to discover new hits and scaffold hopping.
"Molsoft offers excellent tools for computational chemistry. We hope that this synergy will shorten the time of various Hit to Lead projects across the industry." - Robert Kiss, CEO of Mcule
"We are excited to collaborate with Mcule to make 100M potential drug candidates available in the ICM drug discovery suite. It gives our customers another fast way to discover novel chemistry and quickly get on the path to patentable chemicals. The interface inside ICM makes it easy for our customers to search and acquire new lead chemicals via Mcule's compound sourcing services." Andrew Orry Ph.D. Molsoft LLC
About Mcule: Mcule is a chemical marketplace for drug discovery with services based around small-molecule compound sourcing. While the traditional Mcule database aggregates supplier catalogs to cover the in stock chemical space, ULTIMATE contains 122 million novel, synthetically accessible compounds. Mcule recently launched its custom synthesis auction site SynthAgora to cover chemical space not present in any catalogs.
About Molsoft: Molsoft is a leading provider of software tools, databases and consulting services in the area of molecular modeling, cheminformatics and rational drug design. MolSoft offers complete solutions customized for a biotechnology or pharmaceutical company in the areas of computational biology and chemistry. ICM Chemist is a standalone suite of cheminformatics programs for chemical drawing and editing, chemical database generation, chemical searching, clustering, and enumeration. ICM-Pro is MolSoft's main desktop modeling software which can incorporate the tools in ICM-Chemist as well as molecular modeling, virtual screening, and machine learning drug target and ADMET models (MolScreen).
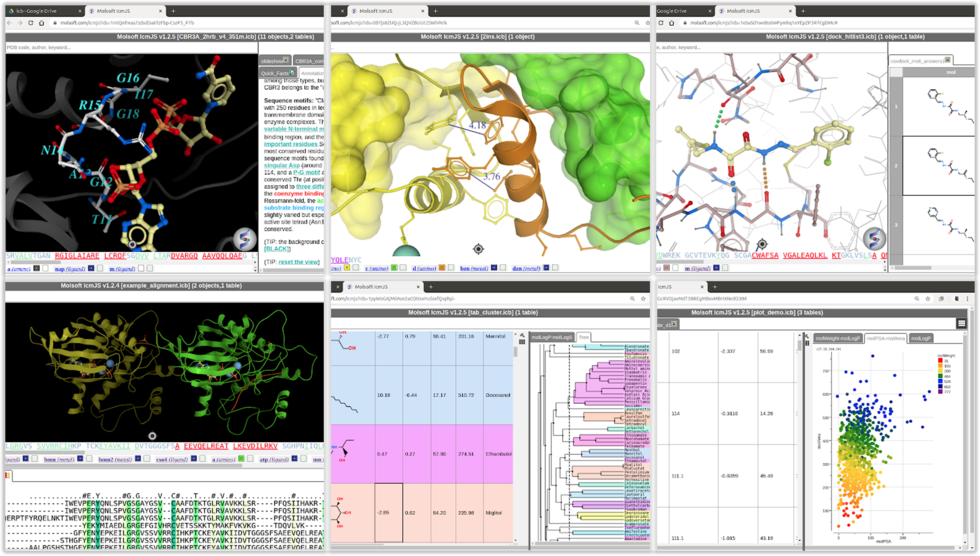 MolSoft is excited to announce the availability of new ICM Java Script (IcmJS) functionality and a convenient add-on for Google Drive. IcmJS is a JavaScript version of MolSoft's ICM Desktop Modeling software and brings the highest quality 3D graphics to the web. IcmJS allows you to view and share with your colleagues fully interactive 3D molecular structures and chemical spreadsheets and plots directly in a web browser. It contains a full suite of molecular visualization tools click here for more information.
MolSoft is excited to announce the availability of new ICM Java Script (IcmJS) functionality and a convenient add-on for Google Drive. IcmJS is a JavaScript version of MolSoft's ICM Desktop Modeling software and brings the highest quality 3D graphics to the web. IcmJS allows you to view and share with your colleagues fully interactive 3D molecular structures and chemical spreadsheets and plots directly in a web browser. It contains a full suite of molecular visualization tools click here for more information.
Here are some examples for you to try: Click here to see some examples using the Google Drive app (download the IcmJS app first).
Arrakis Therapeutics report a new photoaffinity platform for the analysis of chemical interactions with RNA. The paper in ACS Chemical Biology shows how the method they developed called Photoaffinity Evaluation of RNA Ligation-Sequencing (PEARL-seq) can be used for RNA drug discovery by rapidly identifying ligand binding across many types of RNA. MolSoft's ICM PocketFinder method was used to generate an understanding of where small molecules might bind to Aptamer 21 and then ICM docking was used to predict the docked pose of a probe. The docked pose of the ligand was then used to generate hypotheses around which the binding affinity of the ligand could be improved. The SAR data shows that this method has utility for the design and optimization of new RNA-based small molecule therapeutics.
Through this agreement chemists and biologists in India will be able to facilitate access to the software tools and services for lead discovery, ligand docking, screening, modelling, cheminformatics, bioinformatics offered from Molsoft LLC. Under the terms of this agreement Advent Informatics will utilize its marketing and research expertise in India to distribute and support Molsoft products in India.
"High quality modelling tools are very essential in drug discovery, we want to empower the research community in India by providing access to software that is used by leading global pharmaceutical companies" said Surojit Sadhu, Founder, Advent Informatics. "We are excited to enter into this agreement with Advent Informatics because they have a lot of experience in the Indian IT drug discovery market. The ICM-Pro desktop modeling suite is currently outperforming other software for ligand docking and screening accuracy and working with Advent will help expand access to the software for scientists in India said Dr. Andrew Orry (MolSoft LLC).
About Molsoft LLC: Molsoft is a San Diego company that develops new breakthrough technologies in computational chemistry and biology. Molsoft is committed to solving intellectually challenging problems in drug discovery and computational biology. Molsoft has built proprietary computational environments including the ICM suite of programs for molecular modeling, bioinformatics, cheminformatics and ligand docking, and MolCart. Molsoft offers software tools and services in lead discovery, modeling, cheminformatics, bioinformatics, and corporate data management, forms partnerships with biotechnology and pharmaceutical companies, and is the recipient of several NIH and DOE-sponsored awards.
About Advent Informatics: Advent Informatics is a Pune based start-up focusing on providing state-of-the-art software to researchers and students. Advent Informatics promotes solutions from International companies by acting as a bridge between researchers based in India and international software and database companies. Primarily focusing on life sciences, we offer software and IT solutions that catalyse the research of users working in the field of Chemistry, Biology and Drug Discovery, as well as provide technical support offered to its users.
RIDE is a fast 3D molecular similarity search method based on Atomic Property Fields, developed at MolSoft. RIDE searches databases of compound conformers for molecules that are isosteric to the query, i.e. have similar 3D configurations and distributions of atomic properties.
GPU-based implementation is capable of searching ~0.5mln conformers/second on a single GPU card and perform 3D virtual screens of millions of compounds with the level of interactivity comparable to 2D searches. VLS benchmarking on DUDe indicates that RIDE searches produce enrichments on par with much slower standard flexible APF VLS.
RIDE applications include:
A simple web-based interface is provided for the ease of use by chemists, while batch script may be used by chemoinformatitians. In either format, RIDE allows fine-tuning of the queries to generate most desirable hit lists. This includes:
The results of the "blinded" D3R Grand Challenge 4 ligand docking competition have been published and MolSoft once again has outperformed a range of other methods for ligand binding pose and activity prediction.
The work led by Max Totrov Ph.D. (Principal Scientist, MolSoft) has been preented at the D3R evaluation meeting and published in the Journal of Computer Aided Molecular Design (see publication here ). The results from the D3R GC4 are presented along with an analysis of ICM performance in a large (246 complex) macrocycle docking benchmark. Sampling of flexible rings is directly incorporated into the ICM docking and the paper describes the best practices for macrocycle docking.
ICM accurately predicted the pose of BACE macrocycle inhibitors to within sub-angstrom accuracy using multiple receptor and ligand-biased docking. ICM was ranked top in the cognate receptor part of the competition where the organizers provided the protein structure bound to protein. Notably, unlike other top ranking methods, for our submission in this subcategory we have not used any ligand template information.
MolSoft's 4D multiple receptor conformation docking method for incorporating induced fit in the binding pocket was used to dock the macrocycles. The method was improved from GC2 (Lam et al 2018) and GC3 (Lam et al 2018 ) by introducing scoring/energy offsets that optimize the choice of receptor conformation used. In the cross-docking stage of the challenge and when applicable the docking was guided by 3D pharmacophore using MolSoft's Atomic Property Field (APF) method (Totrov 2008).
MolSoft would like to thank the organizers for coordinating the D3R challenge and providing a valuable way of independently assessing the performance of the ICM-Docking and 3D QSAR tools.
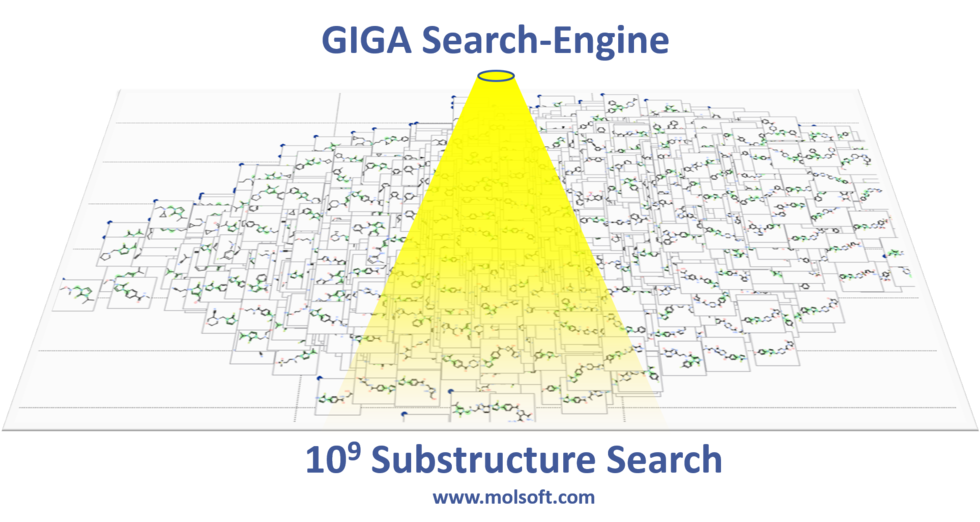 MolSoft is excited to announce the release of MolCart Giga Search - the first of its kind substructure method for efficiently searching databases of 10^9 chemicals in size. Read more about the method here on MolSoft's website here.
MolSoft is excited to announce the release of MolCart Giga Search - the first of its kind substructure method for efficiently searching databases of 10^9 chemicals in size. Read more about the method here on MolSoft's website here.
Databases of synthesizable and virtual chemicals are getting larger each year, therefore the ability to efficiently mine them is critical for drug discovery projects. MolCart Giga Search helps drive your SAR search by providing a way to build target-specific libraries, find derivatives and homologs and screen using SMILES and SMARTs and other chemical properties.
The latest release of MolSoft's ICM-Chemist software provides an easy to use Graphical User Interface for substructure searching of large chemical databases. You can search billions of chemicals or your own custom database using a combination of MolCart Giga Search and ICM-Chemist. Searches can also be scripted and there are large number of post-search processing tools such as Multi Parameter Optimization built into the software.
MolSoft recently collaborated with Enamine to implement MolCart Giga Search on the REAL database (720 million chemicals) you can use it here
Please contact MolSoft for more information about MolCart Giga Search - info@molsoft.com 858-625-2000.
Using ICM for Education - A collaboration between MolSoft and Prof. Charles Grisham at Univeristy of Virginia see publication here: https://t.co/WmdbL97HM4 see the PSAFE resource here: https://t.co/Se968FRgFU
— MolSoft LLC (@MolSoft) March 7, 2019
Thank you very much for your support of MolSoft's webinars. The first webinar series of 2019 attracted more than 650 participants over the course of the five weeks. You can watch the webinars in our Webinar Archives on YouTube or by clicking the links below.
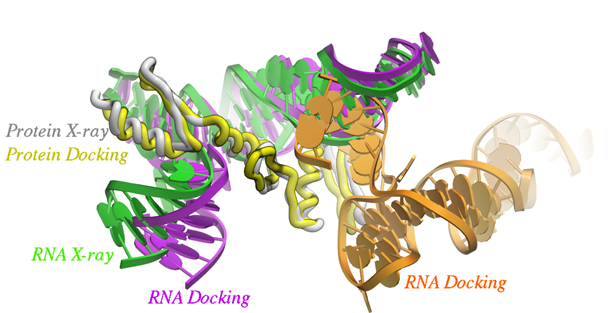 Click here to read paper by Arnautova et al in J Chem Theory Comput. 2018. Paper Abstract: Protein-RNA interactions play an important role in many biological processes. Computational methods such as docking have been developed to complement existing biophysical and structural biology techniques. Computational prediction of protein-RNA complex structures includes two steps: generating candidate structures from the individual protein and RNA parts and scoring the generated poses to pick out the correct one. In this work, we considered three recently developed data sets of protein-RNA complexes to evaluate and improve the performance of the FFT-based rigid-body docking algorithm implemented in the ICM package. An electrostatic term describing interactions between negatively charged phosphate groups and positively charged protein residues was added to the energy function used during the docking step to take into account the greater role that electrostatic interactions play in protein-RNA complexes. Next, the docking results were used to optimize a scoring function including van der Waals, electrostatic, and solvation terms. This optimization yielded a much smaller weight for the solvation term indicating that solvation energy may be less important for the scoring of protein-RNA structures. Rescoring of the generated poses with the new scoring function led to much higher success rates, while pose clustering by contact fingerprints produced further improvements, achieving a success rate of 0.66 for the top 100 structures.
Click here to read paper by Arnautova et al in J Chem Theory Comput. 2018. Paper Abstract: Protein-RNA interactions play an important role in many biological processes. Computational methods such as docking have been developed to complement existing biophysical and structural biology techniques. Computational prediction of protein-RNA complex structures includes two steps: generating candidate structures from the individual protein and RNA parts and scoring the generated poses to pick out the correct one. In this work, we considered three recently developed data sets of protein-RNA complexes to evaluate and improve the performance of the FFT-based rigid-body docking algorithm implemented in the ICM package. An electrostatic term describing interactions between negatively charged phosphate groups and positively charged protein residues was added to the energy function used during the docking step to take into account the greater role that electrostatic interactions play in protein-RNA complexes. Next, the docking results were used to optimize a scoring function including van der Waals, electrostatic, and solvation terms. This optimization yielded a much smaller weight for the solvation term indicating that solvation energy may be less important for the scoring of protein-RNA structures. Rescoring of the generated poses with the new scoring function led to much higher success rates, while pose clustering by contact fingerprints produced further improvements, achieving a success rate of 0.66 for the top 100 structures.
 Scientists at MolSoft led by Max Totrov Ph.D. participated in the Drug Design Data Resource (D3R) Grand Challenge 3 (GC3) and once again achieved great success in the accuracy of docking pose prediction and ligand binding affinity. The competition is a "blinded" open challenge for the worldwide computational chemistry community bringing together teams using different docking methods and software.
Scientists at MolSoft led by Max Totrov Ph.D. participated in the Drug Design Data Resource (D3R) Grand Challenge 3 (GC3) and once again achieved great success in the accuracy of docking pose prediction and ligand binding affinity. The competition is a "blinded" open challenge for the worldwide computational chemistry community bringing together teams using different docking methods and software.
The results and methodology used by MolSoft in GC3 are published in a recent article by Lam et al entitled "Hybrid receptor structure/ligand-based docking and activity prediction in ICM: development and evaluation in D3R Grand Challenge 3". Also, you can watch a video recording of Max Totrov's presentation on the results at the D3R workshop here.
MolSoft's submissions ranked first place for docking pose and affinity prediction for all targets according to key criteria and builds upon MolSoft's successful performance in Grand Challenge 2 (GC2) - see Lam et al J Comput Aided Mol Des. 2018 32:187 "Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach"). In GC2 Molsoft ranked first place for docking accuracy and binding energy prediction.
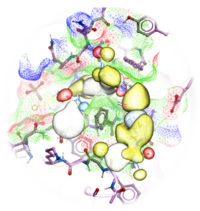
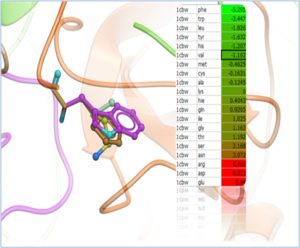
Using MolSoft's ICM-Pro software the team participated in pose and ligand affinity prediction challenges for six different targets including Cathepsin S and five different kinases. The predictions were made using a combination of ICM-docking biased by the Atomic Property Fields (APF) of known ligands acting as templates to guide the pose prediction. The image on the right shows the APF fields for ligand bias for Cathepsin S. This approach provides a way of combining a physics-based docking score as well as a phenomenological APF pseudo energy score for pose ranking. For pose prediction ICM returned a median RMSD for the top pose of 1.7≈ and a median RMSD of 1.06≈ for best pose of five (Figure 1). The disparity between the average RMSD for top pose and the average RMSD for the best pose of 5 is most likely due to crystal contact effects which were not taken into account in the analysis of the results.
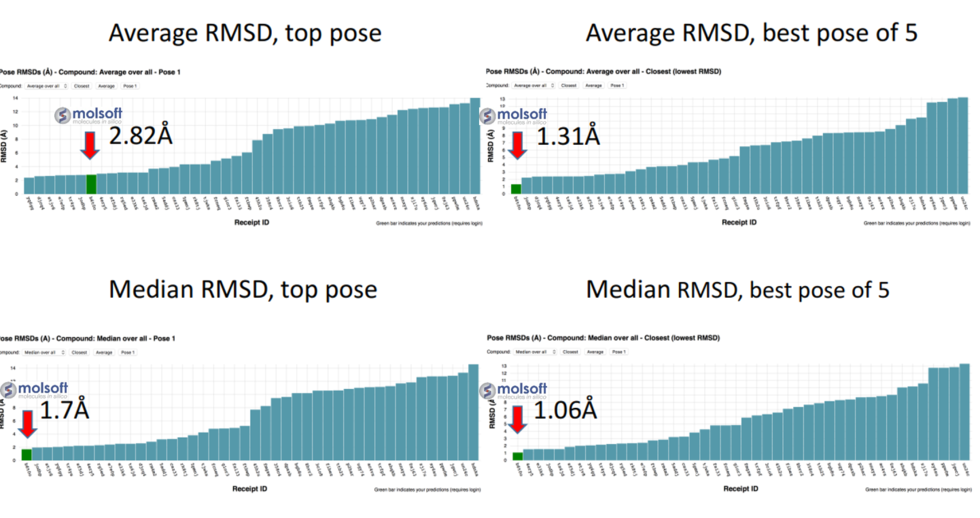 Figure 1: MolSoft's performance in the Pose Prediction challenge of GC3 is highlighted by a red arrow.
Figure 1: MolSoft's performance in the Pose Prediction challenge of GC3 is highlighted by a red arrow.
For the affinity prediction targets it was important to incorporate flexibility of the binding pocket. This was achieved by using pre-aligned PDB structures for each target from the Pocketome database and then selecting up to 10 representative X-ray structures along with compatible ligands used as APF templates.
In the affinity prediction challenge the docked poses of the ligands were scored using a hybrid approach, machine learning (PLS and RFR) on a combination of Receptor/Physics based terms and Ligand/APF-based 3D chemical similarity terms (APF/P 3D QSAR). Evaluated by Kendall tau ranking correlation across all ligands, ICM ranked first place in affinity prediction for all targets (Figure 2).
Figure 2: MolSoft's performance for four targets for the prediction of ligand affinity. Red arrow points at Molsoft's submission.
Resources:
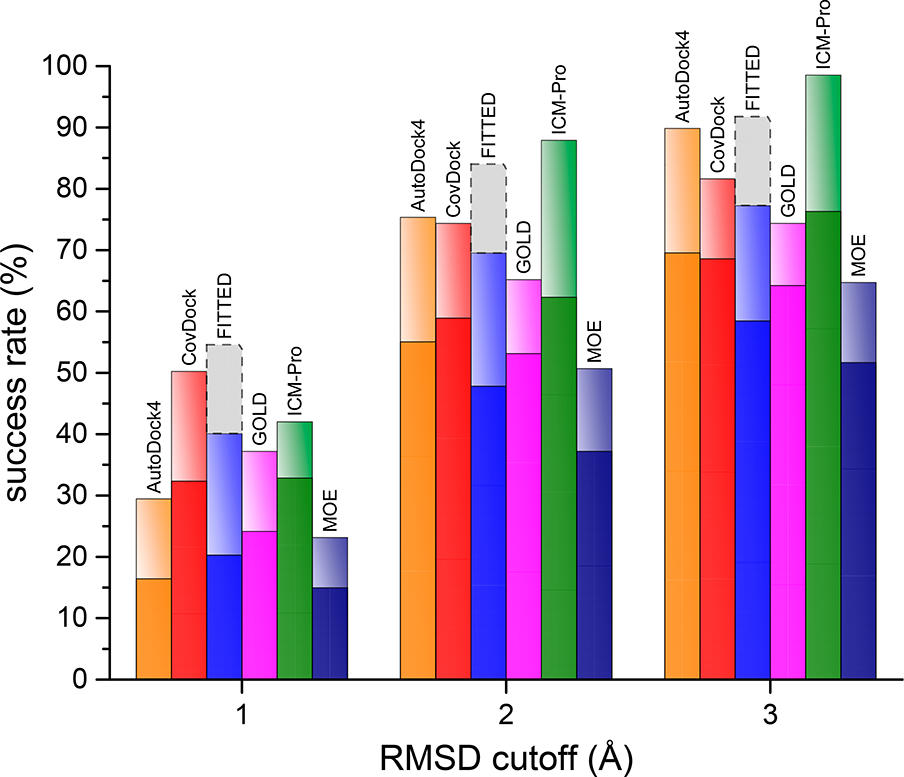
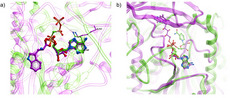
A recent publication by Scarpino et al compared the performance of six covalent docking methods. They selected a large and divergent set of covalent complexes and assessed how well each software could achieve the crystal structure pose of the covalent ligand.
At a two Angstrom RMSD cut off MolSoft's ICM-Pro software was able to achieve the experimental ligand pose in the Top 1 conformations in 62% of the cases and in the Top 10 conformtions in 88% of the cases. MolSoft's ICM-Pro software outperformed the other programs in all tests including reproducing the experimental binding modes and scoring. ICM also performed well with more challenging flexible ligands with 35 heavy atoms or more. In an interesting side test described in the paper, it was found that ICM-Pro's non-covalent conventional docking procedure also gives a remarkable rate of accurate predictions for covalent ligands to modified receptors (CYS/ALA mutations- 86% near native).
The figure below (from Scarpino et al) shows the re-docking success rate at top 1 pose (dark colors) and top 10 poses (lighter colors).

Max Totrov (MolSoft LLC) will present at the "Trends in Physical Properties of Drugs" short course at the Drug Discovery Chemistry Conference in San Diego on Apil 4th. The course topics include:
 Today, Maxm Totrov Ph.D. will present the results of MolSoft's excellent performance in the docking D3R docking challenge at the D3R Workshop. MolSoft's ICM software ranked in first place for average RMSD docking accuracy in the 2016-17 Drug Design challenge competition http://drugdesigndata.org/ (D3R). This work was recently published in the Journal of Computer Aided Molecular Design (see Lam, Abagyan and Totrov 2018).
Today, Maxm Totrov Ph.D. will present the results of MolSoft's excellent performance in the docking D3R docking challenge at the D3R Workshop. MolSoft's ICM software ranked in first place for average RMSD docking accuracy in the 2016-17 Drug Design challenge competition http://drugdesigndata.org/ (D3R). This work was recently published in the Journal of Computer Aided Molecular Design (see Lam, Abagyan and Totrov 2018).
Totrov: Atomic property fields. pic.twitter.com/2tbk47X9Ng
— John Chodera (@jchodera) February 22, 2018
Totrov: Abagyan group has a useful tool called Pocketome that can retrieve and score X-ray structures consistent with ligands. #D3R2018 pic.twitter.com/JfGiSUu3hS
— John Chodera (@jchodera) February 22, 2018
Totrov also points out role of crystal packing contacts in inducing pose flips. pic.twitter.com/uYwdd2aNXs
— John Chodera (@jchodera) February 22, 2018
MolSoft's ICM graphics feature in Chemical and Engineering news article RNA Drug Hunters the https://t.co/t9vlawuQQc Read more about RNA modeling and Lead Discovery using ICM here https://t.co/EhsEsB1EIG #rna #drugdesign #compchem pic.twitter.com/rmWAf0CLz5
— MolSoft LLC (@MolSoft) November 29, 2017
Ruben Abagyan on expanding the drug-target matrix #SBDD2017 pic.twitter.com/6gw7ztzYcu
— Marco De Vivo (@devivo_marco) September 7, 2017
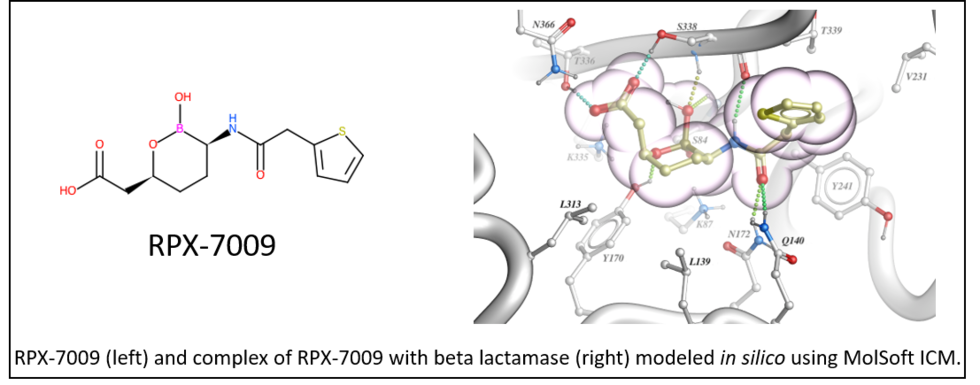
On August 29th 2017 the U.S. Food and Drug Administration (FDA) granted approval of Vabomere to RemPex Pharmaceuticals (part of the Medicines Company ). Vabomere is a new antibacterial drug for the treatment of complicated urinary tract infections which is a life threatening condition (click here for FDA Press Release).
Vabomere is a combination of meropenem (previously developed carbapenem antibiotic) and vaborbactam (RPX-7009), a novel cyclic boronate beta-lactamase inhibitor. Structure-based design by MolSoft's scientists (lead by Maxim Totrov Ph.D.) using the ICM-Pro desktop modeling software was key to the discovery of RPX-7009. The novel cyclic boronate chemotype was first modeled in-silico at Molsoft and prioritized for synthesis based on modeling and docking into several beta-lactamase target enzymes.
The work leading up to this discovery was published by RemPex and MolSoft in the Journal of Medicinal Chemistry - you can read more here.
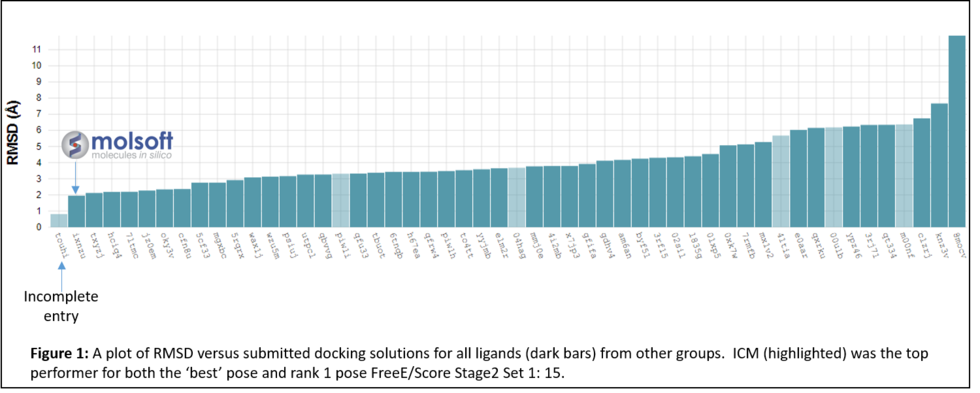
MolSoft's ICM software ranked in first place for average RMSD docking accuracy in the 2016-17 Drug Design challenge competition http://drugdesigndata.org/ (D3R).
Along with over 50 other participants, the Molsoft group led by Maxim Totrov Ph.D.(Principal Scientist, MolSoft), submitted blind docking pose predictions for the Farnesoid X receptor (FXR) which is a drug target for dyslipidemia and diabetes. The MolSoft team used the pocketome (www.pocketome.org) entry for FXR and the ICM-VLS, Atomic Property Fields (APF) and machine learning methods in the ICM-Pro software to predict the interaction of 36 FXR ligands and the binding affinity of 102 other ligands.
In January 2017 the organizers (D3R) distributed the evaluation results which showed that MolSoft's submission ranked first in RMSD for the top scoring pose and was the only one with average RMSD below 2.0A (Figure 1 below). In the Binding Energy Prediction competition ICM APF dock and icm-MMGBSA method ranked in first place and produced the lowest RMSE for Stage 2 Set 1 set of Kd values for 15 ligands (Lam et al 2018).
In Silico Lead Discovery - Free Webinar Series. Register here: https://t.co/SD3DX7eFdh #sbdd #qsar #molecularmodeling #drugdiscovery pic.twitter.com/24CsWVnLub
— MolSoft LLC (@MolSoft) August 25, 2017
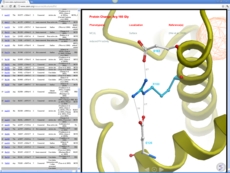
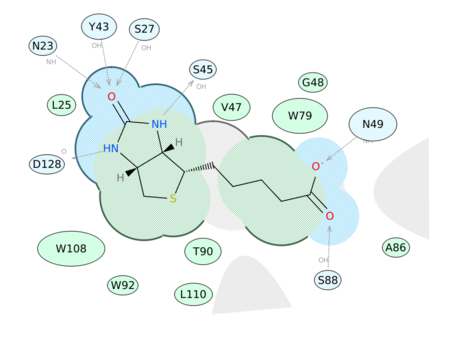 Now you can generate a 2D interaction plot of a ligand with the binding pocket (see image on the right). The image is annotated with hydrogen bonds interacting residues. The residue interactions surface and proximity are represented by the size of the residue label and distance respectively. Grey parabolas and broken thick lines indicate solvent accessible regions and the ligand is shaded by property. [Documentation]
Now you can generate a 2D interaction plot of a ligand with the binding pocket (see image on the right). The image is annotated with hydrogen bonds interacting residues. The residue interactions surface and proximity are represented by the size of the residue label and distance respectively. Grey parabolas and broken thick lines indicate solvent accessible regions and the ligand is shaded by property. [Documentation]
A guide to the coloring and representation of the 2D diagrams:
|
The Macromolecular Transmission Format (MMTF) is a new compact binary format to transmit and store biomolecular structural data quickly and accurately. MMTF is now supported in the latest ICM version it does not have any limitation on number of atoms and is easier to use than the original PDB. You can read more about MMTF here at the PDB website.
The image on the left shows the entire HIV-1 capsid (~2.4 million atoms PDB:3j3q). In ICM (v3.8-5 and higher) use the command below to read in and display a mmtf file.
read pdb mmtf |
 IcmJS (formerly known as ActiveIcmJS) is a JavaScript/HTML5 viewer for 3D Molecular Graphics which does not require any plugin or installation. It runs on all modern browsers including Chrome, Firefox and Safari and is also mobile device friendly. IcmJS gives you full access to the ICM shell and graphics on a web browser. This means that commands available in the free ICM-Browser are also available on the web via IcmJS.
You can view some IcmJS examples here:
IcmJS (formerly known as ActiveIcmJS) is a JavaScript/HTML5 viewer for 3D Molecular Graphics which does not require any plugin or installation. It runs on all modern browsers including Chrome, Firefox and Safari and is also mobile device friendly. IcmJS gives you full access to the ICM shell and graphics on a web browser. This means that commands available in the free ICM-Browser are also available on the web via IcmJS.
You can view some IcmJS examples here:
ActiveICM Reference: Raush, E., Totrov, M., Marsden, B. D. & Abagyan, R. A new method for publishing three-dimensional content. PloS One 4, e7394 (2009).
Want to Learn More? Read more on our IcmJS webpage or register for the IcmJS Webinar here.
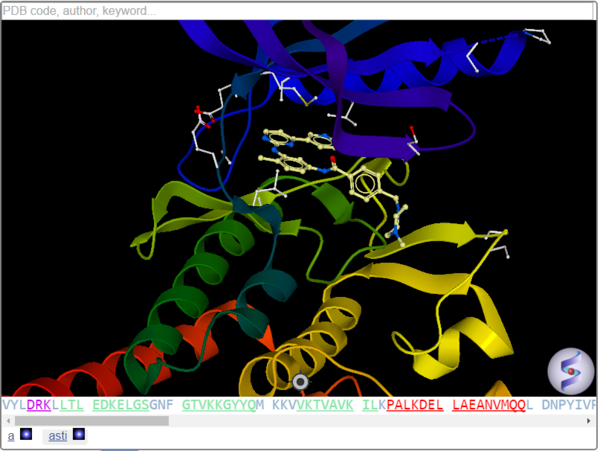
 Prof. Ruben Abagyan (MolSoft Founder and UCSD) will present at talk at the ACS meeting entitled "Toward predictive structural polypharmacology via flexible docking of ligands to the organismal pocketomes".
Prof. Ruben Abagyan (MolSoft Founder and UCSD) will present at talk at the ACS meeting entitled "Toward predictive structural polypharmacology via flexible docking of ligands to the organismal pocketomes".
Please join us for MolSoft's ICM User Group Meeting 2016 to be held in San Diego on March 17-18. Everyone is welcome to attend and we have exciting speakers from academia and the biotech/pharma industry. You can view the meeting schedule here.
Date: March 17-18 2016
Location: MolSoft 11199 Sorrento Valley Road, S209 San Diego CA 92121 [ map ] [ hotels ]
Registration: $99 click here to download a registration form or call 858-625-2000 x108.
Schedule: Click here for full meeting agenda (subject to change)
UGM Archives: See photos and content from previous User Group Meetings here.
Speaker topics will include:
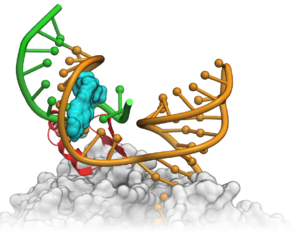 A new paper in Nature Chemical Biology describes the discovery of a potent small molecule which has the potential to be used to treat Spinal Muscular Atrophy (SMA) a debilitating motor neuron disease. The disease is caused by a deficiency of Survival of Motor Neuron (SMN) gene which is the most common cause of mortality in children. The pyridazine class of compounds described in this paper enhance SMN2 splicing which elevates full-length SMN protein. It is proposed that the small molecule modulators enhance binding of U1 snRNP to the SMN2 exon 7 5'ss.
A new paper in Nature Chemical Biology describes the discovery of a potent small molecule which has the potential to be used to treat Spinal Muscular Atrophy (SMA) a debilitating motor neuron disease. The disease is caused by a deficiency of Survival of Motor Neuron (SMN) gene which is the most common cause of mortality in children. The pyridazine class of compounds described in this paper enhance SMN2 splicing which elevates full-length SMN protein. It is proposed that the small molecule modulators enhance binding of U1 snRNP to the SMN2 exon 7 5'ss.
Using MolSoft's ICM Alibero method , scientists at Novartis Institute of Biomedical Research were able to model the binding mode of the molecule with RNA (See Figure- left). To model the flexibility of RNA a population of 150 RNA models was generated and the Alibero method was able to determine the model that best separated known binders from non-binders. On the basis of this model they were able to hypothesize that the small molecules enhance the stabilization of the RNA duplex in the 1A bulge major groove.
FOCUS is the result of more than eight years of collaboration between MolSoft and Novartis. Globally at Novartis there are around 1500 registered users and it has become a popular tool for medicinal and computational chemists as well as structural biologists and bioinformaticians. The core functions of FOCUS can be found in MolSoft's ICM-Chemist-Pro desktop modeling and cheminformatics software product (http://www.molsoft.com/icm-chemist-pro.html). FOCUS contains a wide range of applications including a comprehensive set of feature-rich cheminformatics tools and a 3D ligand editing and design hub. Using MolSoft's ICM scripting language Novartis created customized interfaces that enabled them to build fully interactive html pages, menus, dialogs, panels and table actions all linked to internal and external service calls. The FOCUS development team paid particular attention to make it user-friendly for non-computational researchers.
The Novartis FOCUS development team worked closely with MolSoft's software engineers to build a chemically intuitive and user friendly ligand design interface called the ICM Ligand Editor. The Editor is integrated into FOCUS as well as MolSoft's ICM-Pro and ICM-Chemist-Pro desktop modeling and cheminformatics products. Using the editor, a ligand bound to a protein can be modified in 2D or 3D and the effects of the modification on binding can be immediately calculated and observed. Modifications are stored in the memory, which enables full undo and redo capabilities and the medicinal chemist can conveniently store new designs in fully interactive 3D spreadsheets. Rich graphics tools are available for the representation of the binding pocket surface and interactions between the receptor and ligand. The editor is powered by MolSoft's highly acclaimed ICM docking software and contains options for constrained, induced fit, fragment, and covalent docking. The ligand editor can also be used for 3D pharmacophore modeling using MolSoft's Atomic Property Fields.
The publication describes the development steps that Novartis undertook in the development of FOCUS along with advice for people who want to build a similar platform for their company. If you are interested in finding out more please contact MolSoft (info@molsoft.com) or call 858-625-2000 x108.
 MolSoft is excited to announce that we will host the San Diego leg of the ChEMBL USA tour on January 28th 2015. Please RSVP here.
MolSoft is excited to announce that we will host the San Diego leg of the ChEMBL USA tour on January 28th 2015. Please RSVP here.
About the Workshop: Every year, research produces a vast amount of data describing the biological effects of chemical substances. This valuable information, while public, is usually in a form not accessible for systematic data extraction (data mining) and lacks consistent standardization.
ChEMBL is an Open Data database that contains this information manually extracted from the primary scientific literature. The database contains binding, functional and ADMET information for a large number of drug-like bioactive compounds. Data is further curated and standardized (assay read outs and chemical structures) to maximize their quality and utility across a wide range of chemical biology and drug-discovery research problems.
We have recently complemented ChEMBL with SureChEMBL, a web-based daily updated database of full patent text and automatically extracted compound structures. This allows advanced chemistry based querying of the patent literature. We will present an overview of both resources, and then show specific use common usage and data analysis scenarios
Keynotes Dr. Abagyan @UCSanDiego, Dr. Lipinski @ruleof5guy, Dr. Walters @VertexPharma, and Dr. Brown @Merck. pic.twitter.com/D68TLzR43F
— In Silico DDC (@insiliconf) December 12, 2014
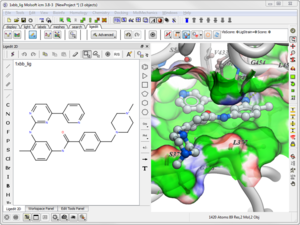
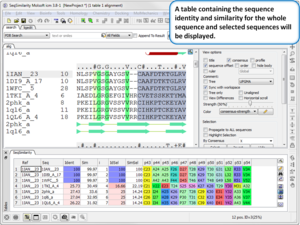 You can download the new version of ICM from our Support Site.
You can download the new version of ICM from our Support Site.
New features include:
Modeling
Sequence and Alignment
Cheminformatics
 MERSi is a new algorithm available in ICM-Pro that allows you to predict the effect of a mutation upon protein stability and protein-protein binding free energy.
MERSi is a new algorithm available in ICM-Pro that allows you to predict the effect of a mutation upon protein stability and protein-protein binding free energy.
Predict Change in Binding Free Energy upon Mutation The binding free energy change ΔΔGbind, is computed as a difference between the binding free energy of mutant and wild type complexes. To account for structure relaxation upon mutation, side chain re-packing in the vicinity of the mutated residue by a Monte Carlo simulations carried out. "Scan Protein Interface" option allows to mutate all residues (one by one) of the Interacting Part in close contact with the second part of the complex. Documentation | Tutorial
Predicting Change in Protein Stability upon Mutation The stability change is calculated as a weighted sum of a complete set of physically meaningful free-energy contributions, including van der Waals attractions, repulsions, electrostatics, hydrogen bonding, and solvation term. Residue-specific constants that account for the free energies of the unfolded and misfolded states were derived empirically using a large set of experimental data. Mutation of a given residue is followed by Monte Carlo simulations with flexible side chains for the mutated residue and its neighboring residues. Backbone flexibility is also considered for special cases such as proline and disulfide-forming/breaking cystein mutations. Documentation
iMolview came out on top in a recent comprehensive review of 12 molecule viewing apps for iPad and Android devices. The review article called "High-Quality Macromolecular Graphics on Mobile Devices: A Quick Starter.s Guide" was published in the Methods and Molecular Biology series by Drs. Yiu and Chen from King's College London, UK. Please click here to download the paper (subscription maybe required).
The article states that iMolview "tops the list because of its user friendliness and it offers the most complete set of functions for visual communication". The paper also includes a useful iMolview tutorial which takes you through the steps of importing molecules, coloring, transparency, surfaces, and exporting images.
You can download iMolview from the Google Play Store. or the Apple App Store.
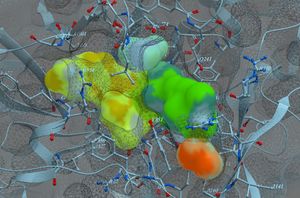
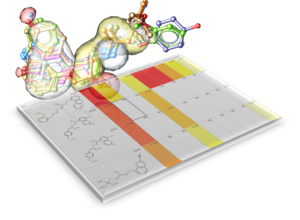
 MolScreen is a set of high quality 2D fingerprint and 3D pharmacophore models for a broad range of pharmacology and toxicology targets. The models can be used for lead discovery or counter screening. The models use MolSoft's 2D QSAR/Fingerprint and 3D Atomic Property Fields ( Totrov 2008 ) methods.
MolScreen is a set of high quality 2D fingerprint and 3D pharmacophore models for a broad range of pharmacology and toxicology targets. The models can be used for lead discovery or counter screening. The models use MolSoft's 2D QSAR/Fingerprint and 3D Atomic Property Fields ( Totrov 2008 ) methods.
The models can be screened directly using MolSoft's ICM-Pro + VLS software. Alternatively we can screen a set of chemicals for you via our contract research services. Please contact us for more information about how to use MolScreen.
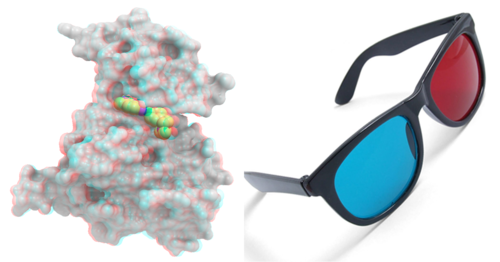
|
MolSoft is excited to announce that Anaglyph 3D is now available to use in ICM and iMolview. Anaglyph 3D can be viewed using low cost 3D glasses and without any expensive 3D compatible hardware or monitors. It can be viewed directly on your computer's monitor, conference room projector, or on your iPad or android device. You can also print the image out rendered for viewing in 3D.
Here are the glasses the developers and scientists at MolSoft are using but any Red and Cyan 3D glasses will work. To use Anaglyph 3D in ICM you will need the very latest version of ICM 3.7-3c and the documentation is here. Please contact us if you do not have the latest version yet. For mobile and tablet users please download the next iMolview update. |

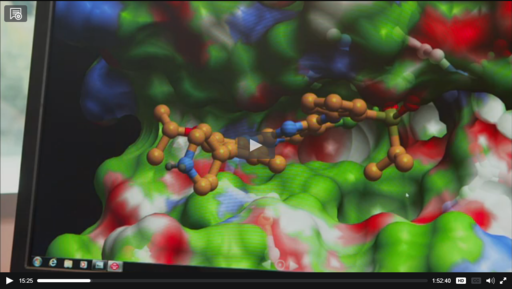
 At the recent Amazon Cloud Computing conference ( AWS Re:Invent ) scientists at Novartis reported using MolSoft's ICM-VLS in the largest ever virtual screen. MolSoft's software developers built a cloud-version of ICM-VLS which enabled Novartis to undertake this ground breaking achievement in cloud-based virtual screening. Cyclecomputing (a cloud vendor service company) enabled the jobs to be distributed to more than 30,000 cores in the USA and Europe. Novartis undertook 10.6 years of drug compound computations in 9 hours! The virtual screen resulted in three new lead compounds which are now being optimized by Novartis. This work was previously reported at MolSoft's user group meeting earlier this year and at BioIT 2012.
Watch a Video - click on image on right.
At the recent Amazon Cloud Computing conference ( AWS Re:Invent ) scientists at Novartis reported using MolSoft's ICM-VLS in the largest ever virtual screen. MolSoft's software developers built a cloud-version of ICM-VLS which enabled Novartis to undertake this ground breaking achievement in cloud-based virtual screening. Cyclecomputing (a cloud vendor service company) enabled the jobs to be distributed to more than 30,000 cores in the USA and Europe. Novartis undertook 10.6 years of drug compound computations in 9 hours! The virtual screen resulted in three new lead compounds which are now being optimized by Novartis. This work was previously reported at MolSoft's user group meeting earlier this year and at BioIT 2012.
Watch a Video - click on image on right.
MolSoft's ICM-VLS has a strong track record of achieving high quality leads in ligand- and structure-based drug discovery. Please contact MolSoft if you would like to learn how you can undertake a similar screen.

MolSoft is excited to announce that they have won a competitive bid to work with AMG and TB Alliance on the development of vital new medicines that can be used to treat Tuberculosis (TB).
TB is a common disease which typically affects the lungs and if not treated correctly can be fatal. The current TB treatment is very long and can result in the patient not adhering to the strict regimen. With the recent approval of Bedaquiline as a treatment for multi-drug-resistant TB it gives hope that new and safe TB drugs can be identified.
In this collaboration, MolSoft will develop ligand-based models using published TB drug activity data as well as internal TB Alliance high throughput screening results. All work will be performed using MolSoft.s Internal Coordinate Mechanics (ICM) molecular modeling and cheminformatics software package which is used worldwide by academic and commercial laboratories. MolSoft will develop ICM 2D fingerprint and 3D Atomic Property Field (APF) models for each of the activity series. The 2D approach uses MolSoft.s proprietary method for generating hashed chemical fingerprints used in chemical search and similarity calculations. The 3D APF method, developed by Molsoft.s Principal Scientist Dr. Maxim Totrov, is a 3D pharmacophoric potential implemented on a continuously distributed grid which can be used for ligand docking and scoring.
Dr. Ruben Abagyan (MolSoft.s Founder and Professor at Skaggs School of Pharmacy and Pharmaceutical Sciences UCSD) says "The collaboration between chemists at TB Alliance and the scientific team at MolSoft represents an exciting application of their ICM ligand-based modeling and screening methods. MolSoft.s scaffold hopping technology is being used successfully in the pharmaceutical industry and it is a sign of this achievement that MolSoft was chosen to work on this project. I am confident that this alliance will give the best chance of identifying a new class of TB drugs."
Once the models have been prepared ICM-VLS will screen >12 million drug-like compounds to each model. High quality novel hits will then be chosen which are suitable according to the project.s pharmacokinetic and cytotoxicity criteria for hit to lead progression.
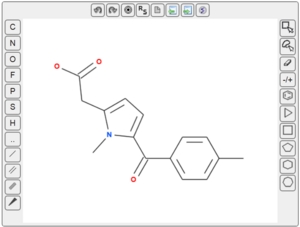
MolSoft announces the release of a HTML5 Molecular Editor which allows you to draw 2D chemical structure inside the web browser's page. It does not require any plug-ins (like Java) and works in any modern HTML5 compatible browser with Java Script enabled (including mobile platforms). You can sketch chemicals as well as import and export in MOL or SMILES format.
The editor is made available free of charge to academic institutions, please contact us if you wish to use it for commercial use. Please click here for more information on how to use the editor in your application.
To see the HTML5 Molecular Editor in action please visit one of our free popular online services.
iMolview v1.7 iPhone/iPad
Tools/Load Electron Density Map
Loads electron density map from Uppsala EDS server (http://eds.mbc.uu.se/eds/). Once map is loaded you can toggle its display using 'Misc' panel in the main menu. In addition map can be contoured around selected atoms:
New icons in 'Misc' panel:
iMolview v1.2 Android
The key new features are: Release Version 1.1

Thank you very much to all the participants of our workshop entitled "April 6th 2013: Next generation of ICM desktop chemistry and biology modeling tools Workshop 2013". It was an exciting workshop highlighting all the new features of ICM and a half day user group session. Please click here to view the contents of the meeting and the speaker abstracts.
The new features include:
 MolSoft has released its popular iMolview mobile software for the Android today. Please download iMolview from the Android version from the Google Play Store..
MolSoft has released its popular iMolview mobile software for the Android today. Please download iMolview from the Android version from the Google Play Store..
About iMolview Explore the fascinating world of biological macromolecules: iMolview lets you browse and view in 3D protein and DNA structures from Protein Data Bank. Search for proteins like 'insulin' or 'thyroid receptor' in PDB or simply type pdb code the search bar. Information associated with each molecule in these databases is also at your fingertips. Sync and view your own structure files via iMolview folder on your sdcard. Molecular view can be customized with a rich set of molecular representations (wires, balls-and-sticks, space filling, ribbon diagrams, molecular surfaces) and various coloring schemes. Select residues, atoms or chains and color or change their representations individually. Select 'neighbors' of a ligand or any other selection to identify interacting atoms or residues. Set 'inertia' to the maximum and let your molecule spin in 3D indefinitely.
 |  |
 |
Last year scientists at MolSoft (Max Totrov Ph.D.) and UCSD (Ruben Abagyan Ph.D. and Marco Neves Ph.D.) competed in an industry-wide docking and screening competition organized by OpenEye, Merck, and GSK. Two challenges were set:
Pose Prediction
Virtual Screening Recognition
MolSoft ICM achieved excellent results in both challenges ranking in first place in both challenges. ICM-Pro docking gave pose prediction results of < 1A top 1 pose: 78% of cases and < 2A top 1 pose: 91% of cases. For virtual screening, median area under ROC curves equal to 69% and 82% were achieved. A full description of the data analysis can be read here
UGM 2012 Links
Photos by Dr.Polo Lam - MolSoft and Photos by Eugene Raush - MolSoft

MolSoft is excited to announce the release of ICM version 3.7-2c. The new version can be downloaded from our support site here. The key new features are described below but for a detailed list please see the release notes.
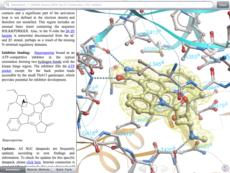
 Atomic Property Fields APF is a 3D pharmacophoric potential implemented on a grid (Totrov 2008 and 2011 ). APF can be generated from one or multiple ligands and seven properties are assigned from empiric physico-chemical components (hydrogen bond donors, acceptors, Sp2 hybridization, lipophilicity, size, electropositive/negative and charge). An independent study found that APF is most accurate method for chemical superposition and ligand-based 3D virtual screening compared to several other commercial and academic softwares. (Giganti et al 2010).
Atomic Property Fields APF is a 3D pharmacophoric potential implemented on a grid (Totrov 2008 and 2011 ). APF can be generated from one or multiple ligands and seven properties are assigned from empiric physico-chemical components (hydrogen bond donors, acceptors, Sp2 hybridization, lipophilicity, size, electropositive/negative and charge). An independent study found that APF is most accurate method for chemical superposition and ligand-based 3D virtual screening compared to several other commercial and academic softwares. (Giganti et al 2010).
 3D ligand Editor The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor.
3D ligand Editor The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor.
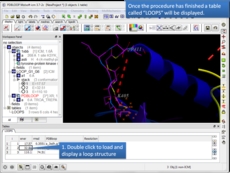
Protein Modeling Inside ICM there are many features for homology modeling and loop modeling. This new option can be used if you have a gap in your protein and you want to find loops in the PDB which fit the gap.
Chemisty A couple of new options in the Chemistry menu.
 "Pipe-able" Scripting in ICM New options to pipe icm commands and scripts.
"Pipe-able" Scripting in ICM New options to pipe icm commands and scripts.
Other New Features and Options
 | Today MolSoft released a new version (v1.4) of its popular molecular viewing app called iMolview for the iPad and iPhone. The new features include the ability to save molecular documents which include structure and slides. A slide is a combination of a view point and graphics representation. In iMolview you can create multiple slides and access them later after the document was saved.
To create a slide:
To switch between slides you may use Menu/Slides or arrows on the sides of the center button. After all slides are done you need to save your molecular document. Goto Menu/Documents/Save Current Document and give it a name. The document will be saved into History folder and can be loaded later. This file can also be copied to your PC,MAC or Linux and opened with free ICM-Browser, or you can embed this document as a live 3D object into web page using ActiveICM technology. iMolview costs only 99cents and is available via the Apple App Store. Please send us your feedback on the new version and rate the app at the Apple app store. |
 | Prof. Abagyan's lab at UCSD have developed a website called the Pocketome which is an encyclopedia of conformational ensembles of all druggable binding sites that can be identified experimentally from co-crystal structures in the Protein Data Bank. The pocketome ensembles are generated from UniProt and PDB and clusters and validates the binding pockets based on the consistency of the composition between multiple structures. The pockets can then be viewed using ActiveICM or downloaded and viewed using ICM-Browser.
|
 |
SummaryThe community-wide GPCR Dock assessment is conducted to evaluate the status of molecular modeling and ligand docking for human G protein-coupled receptors. The present round of the assessment was based on the recent structures of dopamine D3 and CXCR4 chemokine receptors bound to small molecule antagonists and CXCR4 with a synthetic cyclopeptide. Thirty-five groups submitted their receptor-ligand complex structure predictions prior to the release of the crystallographic coordinates. With closely related homology modeling templates, as for dopamine D3 receptor, and with incorporation of biochemical and QSAR data, modern computational techniques predicted complex details with accuracy approaching experimental. In contrast, CXCR4 complexes that had less-characterized interactions and only distant homology to the known GPCR structures still remained very challenging. The assessment results provide guidance for modeling and crystallographic communities in method development and target selection for further expansion of the structural coverage of the GPCR universe. |
 | This month the Journal of Inherited Metablic Disease published a fully interactive 3D research paper using MolSoft's ActiveICM. The paper describes the crystal structure of human fumarate hydratase along with mutations that are mapped onto the 3D structure. The reader can click on a mutation or group of mutations and the fully interactive 3D display takes the reader to the location in the crystal structure in 3D. The reader can then interact (zoom, rotate etc..) with the molecule making the it easier to visualize the data. You can read more on the SGC website here. |
We were excited to read a review of MolSoft's iMolview app for iPad and iPhone on the popular crystallography P212121.com website. Click here to read it... The P212121.com website is not only a blog but also a macromolecular crystallography equipment and reagent store. Scientists can browse thousands of products from dozens of companies all from a single website, this not only saves you time but you also avoid the sales reps! The iMolview app is being used around the world by scientists in many disciplines and we have received many positive reviews. The app is under active development, so if you are a user and there is a feature you would like to see please contact us. If you have not tried iMoview yet you can do so by purchasing it from the apple store for only 99 cents.
 | Max Totrov (Principal Scientist, MolSoft LLC) recently gave a lecture in Korea on "Ligand Binding Site Superposition and Comparison based on Atomic Property Fields". You can watch the lecture here and read more about this in this published paper. |
 | MolSoft LLC were today awarded a patent entitled "Structured documents and systems, methods and computer programs for creating, producing and displaying three dimensional objects and other related information in those structured documents" for its ActievICM technology (US Patent Number 7,880,738). The patent is for a structured document file which includes graphical and non graphical information. The graphical information includes the vector and coordinate description of a representation of at least one three dimensional object and at least one of the following group of view attributes or features: rotation, translation, view angle, lighting, colors, specific graphical representations, labels and parameters. The vector and coordinate description are stored with or independently from the one or more view attributes or features and the vector and coordinate description and the one or more view attributes are stored in a single file which can be accessed and viewed by a structured document viewer. Related authoring tools and viewers are also disclosed. |
 Nature Structural and Molecular Biology is now using using MolSoft's ActiveICM technology. The first paper published in this enhanced format in Nature Structural and Molecular Biology can be viewed here. Thank you to the Editors at Nature, the authors of the paper, and Dr. Brian Marsden and Dr. Wen Hwa Lee at SGC for their support and for making this possible. ActiveICM is developed by MolSoft and allows you to view fully-interactive 3D molecules and animations on the web or PowerPoint.
Nature Structural and Molecular Biology is now using using MolSoft's ActiveICM technology. The first paper published in this enhanced format in Nature Structural and Molecular Biology can be viewed here. Thank you to the Editors at Nature, the authors of the paper, and Dr. Brian Marsden and Dr. Wen Hwa Lee at SGC for their support and for making this possible. ActiveICM is developed by MolSoft and allows you to view fully-interactive 3D molecules and animations on the web or PowerPoint.
Dr. Ruben Abagyan who is Molsoft's founder and Professor at the Skaggs School of Pharmacy at UC San Diego said, "It is great to see ICM and its derivative, activeICM, revolutionizing the way scientific information is communicated to the public. Since their creation 20 and 6 years ago respectively we managed to transform this software into a sophisticated multi-platform environment. We are committed to the continuous development of this platform and making it freely available for the research community."
To view the ActiveICM content:
You can make similar files using the free ICM-Browser http://www.molsoft.com/icm_browser.html
 MolSoft recently developed ChemDiv's new eShop containing the world's largest small molecule inventory. The eShopuses MolSoft's proprietary cheminformatics tools to enable you to search the database by sketching the structure or part of a structure, by ID number, SMILES, or by chemical files such as Mol and SDF. A template structure can also be used to build libraries based around a particular substructure. There are also chemical property filtering tools available such as MW, logP, and Number of rotatable bonds, hydrogen bond donors and acceptors.
MolSoft recently developed ChemDiv's new eShop containing the world's largest small molecule inventory. The eShopuses MolSoft's proprietary cheminformatics tools to enable you to search the database by sketching the structure or part of a structure, by ID number, SMILES, or by chemical files such as Mol and SDF. A template structure can also be used to build libraries based around a particular substructure. There are also chemical property filtering tools available such as MW, logP, and Number of rotatable bonds, hydrogen bond donors and acceptors.
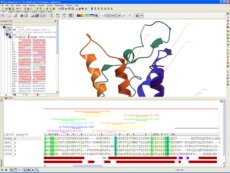
MolSoft is excited to announce the release of ICM v3.7-2 which is a major new version upgrade with many new features and an updated user interface. You can download the latest version at our support site. Click here for more information on the new features available in ICM v 3.7.
New feature hightlights include:

Scientists at MolSoft have developed a new physics-based internal coordinate mechanices force field. Please see the publication in the Proteins journal by Arnautova et al where the force-field has been applied to protein loop modeling. The new force-field contains new parametization for the dielectric constant, an improved hydrogen bond determination method, and implementation of novel backbone atom torsional potentials which include bond anlges of the carbon (alpha) atoms into the internal variable set.
Follow these instructions to use the new force field in your own scripts for energy minimization and optimization. If you are interested in modeling loops you can use the _loopmodel macro where all these parameters are already pre-set.
LIBRARY.res = {"icmff"}
read library
dielConst = 2.
set term "bb"
ffMethod = 4

The development of ActiveICM otherwise known as iSee has been a result of a strong collaboration between scientists at the SGC and developers at MolSoft. If you are attending the Bio-IT World Conference in Hannover next month please be sure to attend the talk by Dr. Brian Marsden from the SGC entitled "iSee - Dissemination of Structural Biology Data to the Masses". The talk will be at 11:30AM on Wednesday 6th October in the IT Infrastructure and Informatics session,please click here to read the abstract for his talk.

Following on from the success PLoS ONE and PLoS Biology have had using ActiveICM it gives us great pleasure to let you know that the Molecular and Cellular Proteomics Journal, published by the American Society for Biochemistry and Molecular Biology (ASBMB) are now accepting articles in ActiveICM format. Two articles have been published in this format already. Please click on the link below to view them. Remember to download the free ActiveICM plugin to view them.
John H. Morris, Elaine C. Meng and Thomas E. Ferrin (2010) Computational Tools for the Interactive Exploration of Proteomic and Structural DataAugust 1, 2010 Molecular & Cellular Proteomics, 9, 1703-1715. : doi: 10.1074/mcp.R000007-MCP201
Keren Lasker, Jeremy L. Phillips, Daniel Russel, Javier Velazquez-Muriel, Dina Schneidman-Duhovny, Elina Tjioe, Ben Webb, Avner Schlessinger and Andrej Sali (2010) Integrative Structure Modeling of Macromolecular Assemblies from Proteomics Data August 1, 2010 Molecular & Cellular Proteomics,9,1689-1702. doi: 10.1074/mcp.R110.000067
 Click to enlarge image.
Click to enlarge image.
This week PLoS Biology have started using MolSoft's ActiveICM software to publish fully interactive 3D papers. This is following on from the recent success of the series in PLoS ONE. The articles allow the user to rotate, translate, and zoom 3D structures whilst reading the scientific paper and view animations directly linked to hyperlinked text. The publication of these papers is due to a collaboration between MolSoft and the SGC. Click on the links below and then choose Enhanced Version to view the latest papers in this fully interactive 3D format. You will need to download ActiveICM to view them.
Rellos P, Pike ACW, Niesen FH, Salah E, Lee WH, et al. (2010) Structure of the CaMKIIG/Calmodulin Complex Reveals the Molecular Mechanism of CaMKII Kinase Activation.PLoS Biol 8(7): e1000426. doi:10.1371/journal.pbio.1000426
Davis TL, Walker JR, Campagna-Slater V, Finerty PJ Jr, Paramanathan R, et al. (2010) Structural and Biochemical Characterization of the Human Cyclophilin Family of Peptidyl-Prolyl I somerases.PLoS Biol 8(7): e1000439. doi:10.1371/journal.pbio.1000439

In a recent publication by Giganti et al (Pasteur Institute, France) in the Journal of Chemical Information and Modeling ICM displayed the best performance in terms of molecular alignment and docking accuracy compared to four other software programs. The authors acknowledge the efficiency of MolSoft's Biased Probability Monte Carlo method which is good at sampling flexibility. You can read about the comparison here. The authors tested ICM-docking and the Atomic Property Field (APF) method. The APF method is described here: Totrov M (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 71: 15-27
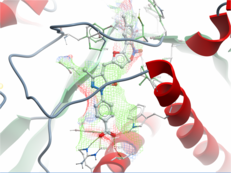 Click to enlarge image.
Click to enlarge image.
A collaboration between scientists at MolSoft and the John Hopkins University has resulted in the discovery of a potent selective small molecule inhibitor of p300/CBP Histone Acetyltransferase (P300 HAT). MolSoft scientists screened ~500K molecules using ICM-VLS and selected 194 for testing which resulted in 30 active compounds. P300 HAT is a transciptional activator implicated in many gene regulatory pathways and protein acetylation and is an important anticancer target. Read more here in the article published in Chemistry and Biology.

MolSoft's ICM-Browser-Pro software was used to make all the molecular graphics images in Dr. Bernhard Rupp's new book called "Biomolecular Crystallography: Principles, Practice, and Applications to Structural Biology" published by Garland Science. To find out more about the book please click here and to read a review of the book please see this article in Acta Crystallographica Section D.
Scientific publishing is now going 3D. For the first time in online publishing history, the PLoS ONE journal is publishing original research papers containing fully annotated and interactive 3D objects. This 3D environment provides a "movie-like" experience but with a significant difference - the reader is in control of the scenes and each view is linked directly to the text. This is possible because of a new technology called ActiveICM developed by MolSoft LLC. ActiveICM is now being used for scientific publishing due to a collaboration with scientists at the SGC. Initially the technology is being applied to the life sciences for the 3D display of the atomic structures of molecules implicated in disease. ActiveICM can also be used for any online publication where a 3D view would enhance the subject matter. Click here to see the PLoS ONE collection of papers. Click here to read the press release.
 Please visit our support site to download the latest MAC version 3.7-1f of ICM-Browser, ICM-Browser-Pro, ICM-Chemist and ICM.
Please visit our support site to download the latest MAC version 3.7-1f of ICM-Browser, ICM-Browser-Pro, ICM-Chemist and ICM.
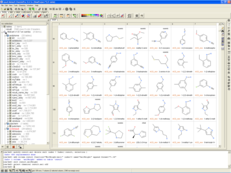 Click to enlarge image.
Click to enlarge image.
The MolCart Building Block database has been updated. The database consists of chemical building blocks for combinatorial chemistry from 32 different chemical vendors and contains 4,408,865 non-redundant chemical fragments. The database contains a wide variety of scaffolds which can be used to generate libraries using Molsoft's combinatorial library tools contained in ICM-Chemist. ICM-Chemist contains a large selection of tools for chemical enumeration and decomposition. You can create or modify a Markush structure and enumerate a library on the fly by adding chemical fragments to defined "R-groups". You can also enumerate by chemical reaction and decompose a database into chemical fragments.
A new 64-bit Beta version of ICM is available. Please contact andyATmolsoft.com if you would like to try this version of ICM.
 Click to enlarge image.
Click to enlarge image.
ICM graphics is featured on the front cover of a new book edited by Edgar Jacoby (Novartis) on Chemogenomics. More information about the book can be found here. The image is of the contoured electron density of a ligand. Click here for instructions on how to contour electron density in ICM.
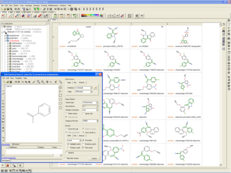 Click to enlarge image.
Click to enlarge image.
The Molcart Compound Screening Database has been updated. The database is a collection of compounds from 33 chemical vendors who let us know whenever a new set of compounds is added or removed from their database.The database currently contains 8,275,223 compounds and the total number of redundant compounds is 4,474,597. The latest changes to the database are documented here. One of the advantages of the MolCart Compound Screening Database is that we take great care to ensure the compounds that are listed are actually available for purchase. Other compound database collections list compounds that are virtual and in theory can be synthesized but they are expensive and are rarely available for quick purchase. If you are a MolCart customer please update your database collection by clicking here and download individual vendor databases or our non-redundant compilation. If you are interested in purchasing MolCart you can get a 2 week trial license by contacting us at info@molsoft.com.
MolSoft LLC (www.molsoft.com) is a leading developer of breakthrough technologies in computational biology and chemistry. MolSoft has officially released a new ground-breaking tool for the interactive, three-dimensional (3D) design and docking of new drug candidates. The ICM Interactive 3D Ligand Editor is an easy-to-use desktop application that allows a chemist to visualize a receptor-ligand complex and then apply their experience and intuition to explore, design and optimize new drug-like compounds. Recently, this tool was validated in a community-wide evaluation of modeling and docking software where ICM performed very successfully in predicting the pose of a ligand to a G-Protein Coupled Receptor (see Michino et al Nature Reviews Drug Discovery 8: 455-456).
A common scenario in drug design is that a lead compound binds reasonably well to a target protein and the ligand-receptor interactions are known, but the problem of how to improve the binding properties, synthetic feasibility, and metabolic-physicochemical properties of the ligand remains. This is where the power of the ICM 3D Ligand Editor comes in: it allows the chemist to see the ligand-receptor interactions clearly on the computer screen. Chemists can simply explore their new ideas with a click of a mouse in a fully interactive and intuitive 3D environment. The effect of a change to the ligand in terms of its interaction with the receptor is immediately displayed on the screen for the chemist's immediate evaluation.
The pharmaceutical industry is already a major user of MolSoft's ICM 3D Ligand Editor technology and industry feedback has enabled MolSoft's developers to build a tool suited for chemists working in drug discovery research. Dr. Eric Martin, Director of Computational Chemistry at the Novartis Institute for Biomedical Research says "The ICM ligand editor is an extremely effective drug design tool for interactively merging the rigor of complex molecular calculations with the intuition of experienced medicinal chemists. We use it extensively both 1-on-1 at the graphics workstation, and in team meetings using a 3D projection system".
So how do you use the ICM 3D Ligand Editor? The process is simple and designed to make life easier for the chemist. First, the ICM 3D Ligand Editor displays the crystal structure or model of the receptor-ligand complex along with the properties of the pocket, ligand solvent accessible surface, ligand strain or unsatisfied hydrogen bonds. The chemist can then start exploring ideas for improving the ligand by selecting a new chemical group and adding it to the ligand. The binding energy and interaction with the receptor is immediately assessed and displayed on the screen. The chemist has access to a full panel of chemical groups, linkers, bond types, stereo/isomer options and atom selections, which can be used to quickly try many different ideas. Importantly, should something be undesirable, a key feature allows the chemist to quickly undo changes to the ligand with a single mouse click and get back to the original ligand. Once the chemist has designed a ligand they are happy with, it can be saved in a single file to share with colleagues. The editor is a perfect tool for communication among scientists and can aid brain-storming session in an exciting way because hypotheses can be tested instantaneously.
The ligand editor has many advanced functions that make use of MolSoft's much acclaimed ICM technology, which is the backbone of the tool. Inside the editor, features such as ligand minimization, docking, atom tethering and virtual screening are available to use. For example, thousands of chemical groups can be screened on a particular point of the ligand resulting in a ranked list of complexes. The editor can also be used for fragment based drug design and optimization. According to Professor Ruben Abagyan, MolSoft Founder and professor at the Scripps Research Institute, "This new interactive environment for lead discovery combines the best practices in flexible docking and interactive modeling. The user-friendly environment changes the way chemical probes are discovered in industry and academia. Furthermore, the freely available ICM Browser and ActiveICM plugin allow scientists to disseminate data among chemists and biologists in an interactive way."
The ICM 3D Ligand Editor is part of the ICM-Chemist-Pro software product available from MolSoft LLC (www.molsoft.com).
A popular request from the biologists who use ICM is to develop a way to visualize sequence conservation in an alignment. Therefore in ICM version 3.6-1e (to be released soon) we have developed a way to display the sequence profile in a multiple sequence alignment. The consensus strength of a residue at a point in the alignment is visually represented by the size of the letter in the profile (see accompanying image).
To display the sequence profile:
Modeling structures of G Protein Coupled Receptors (GPCRs), the most prevalent drug targets, has always been a great challenge. Predicting how and where the drugs bind to those receptors is an even greater challenge. Until recently only bovine rhodopsin and beta 2 adrenergic receptor structures have been solved. Yet, when the structure of the Adenosin receptor A2a (the receptor for caffeine) was solved in a complex with a drug, the crystallographers annouced a challenge for all modellers and dockers to predict the binding interactions of the drug with the receptor.
60 groups have registered for the competition and 29 dared to submit their models. Up to 10 models were allowed to be submitted. More importantly the models were supposed to be ranked since usually only the top predicted pose is further pursued.
In a recent paper by Michino et al published in Nature Reviews the results are published (click here to read the paper). Two teams from Molsoft, Katrich-Abagyan and Lam-Abagyan built the models that had the largest number of correct ligand-receptor interatomic contacts, 45 and 34 out of 70, respecitvely out of all participants. Moreover, both models were ranked number 1, while the prediction from a competing modeler thathad better drug RMSD (when a flexible drug ring was considered) was only ranked 2. The fourth model from the Baker/Rosetta group had only 18 of correct contacts and this pose was only ranked 4th.
The ICM-based GPCR modeling approach employs the Ligand-guided principle and the ICM global optimization algorithm, and its success in ligand docking shows applicability of the method to a large class of problems in the field of GPCR drug discovery.
A new edge accent feature is available in icm version 3.6-1e which will allow you to make images as shown in the pictures below.
The option can be activated in the command line using:
GRAPHICS.sketchAccents = yes
Click on the images below:
 Picture by: Dr.Lars Brive (Goteborg University)
Picture by: Dr.Lars Brive (Goteborg University)
Thank you to everyone who attended the ICM User Group Meeting last month. It was a really enjoyable meeting with some great presentations. Click here to view the meeting program with speaker abstracts and bio.
Thank you to Dr. Lars Brive (Goteborg University) for his photos of the lunch at the beach (small/large) and also to Prof. Bengt Persson (Linkˆping University & Karolinska Institute) whose photos can be viewed here:
 |
| ICM User Group Meeting - Photos Provided by Prof. Persson |
MolSoft's ICM-VLS was used by scientists in Canada and Hong Kong to screen the NCI ligand database to identify a synthetic compound which appears to be able to stop the replication of the influenza viruses, including bird flu virus. See the paper at J Med Chem webiste and here at MSNBC.
Researchers from Virginia Tech and Molsoft LLC have received a five-year, $3.557 million grant from the National Institute of Allergy and Infectious Diseases (NIAID) to continue their promising work on a new class of resistance-breaking insecticides to reduce malaria transmission. Click here for the full story.
 | Thank you to Dr. Kandeel at Gifu University in Japan who let us know that one of the images from his paper in Parasitology was included on the front cover of the January issue of Parasitology. The image was made in ICM-Browser-Pro, see the front cover here. |
MolSoft Newsletter: December 2008 In this issue:

Abstract:Type-II kinase inhibitors represent a class of chemicals that trap their target kinases in an inactive, so-called DFG-out state, occupying a hydrophobic pocket adjacent to the ATP binding site. These compounds are often more specific than those that target active DFG-in kinase conformations. Unfortunately, the discovery of novel type-II scaffolds presents a considerable challenge, partially because the lack of compatible kinase structures makes structure-based methods inapplicable. We present a computational protocol for converting multiple available DFG-in structures of various kinases (?70% of mammalian structural kinome) into accurate and specific models of their type-II bound state. The models, described as deletion-of-loop Asp-Phe-Gly-in (DOLPHIN) kinase models, demonstrate exceptional performance in various inhibitor discovery applications, including compound pose prediction, screening, and in silico activity profiling. Given the abundance of the DFG-in structures, the presented approach opens possibilities for kinome-wide discovery of specific molecules targeting inactive kinase states.

November 2008 Newsletter
Welcome to the November 2008 MolSoft ICM Newsletter containing information on:
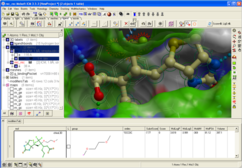 We are pleased to announce the release of two new stand alone Cheminformatics products called ICM Chemist and ICM Chemist Pro. ICM Chemist is a low cost feature-rich product for dealing with chemical structure and databases. The ICM Chemist Pro product contains all the features in ICM Chemist and gives you access to the 3D world of chemical viewing and manipuation. Some of the main features of each of the products are listed below
We are pleased to announce the release of two new stand alone Cheminformatics products called ICM Chemist and ICM Chemist Pro. ICM Chemist is a low cost feature-rich product for dealing with chemical structure and databases. The ICM Chemist Pro product contains all the features in ICM Chemist and gives you access to the 3D world of chemical viewing and manipuation. Some of the main features of each of the products are listed below
ICM Chemist Features
April 9-10 2009 ICM User Group Meeting.
We are looking forward to the 1st ICM User Group Meeting in April next year. The meeting schedule will include talks by experienced ICM users from around the world with an emphasis on how they have successfully used ICM and other applications for their research. MolSoft's developers and scientists will also be presenting at the meeting, so it will offer a good opportunity for you to hear about the latest ICM developments and new features. More information about how to register for the user group meeting and the speakers will be in the next newsletter.
Feb 5-6 2009 - ICM Workshop "Protein Structure and Drug Design"
October 1-2 2009 - ICM Workshop "Protein Structure and Drug Design"
More details here
 The ICM Language Manual and the ICM User Guide Manual can now be google searched. Click here to give it a try. Once you have performed a search you can refine it to only display results from the Language Manual or the User Manual. This search will soon be integrated into the online versions of the manual.
The ICM Language Manual and the ICM User Guide Manual can now be google searched. Click here to give it a try. Once you have performed a search you can refine it to only display results from the Language Manual or the User Manual. This search will soon be integrated into the online versions of the manual.
 Click here to view an example of our new ActiveICM product (US Patent No:7,880,738) in action. The files used in ActiveICM can be generated using ICM Browser Pro, ICM Chemist, or ICM Pro and ActiveICM is free to customers of this product. The online example is a Structural Genomics Consortium datapack file of PAK4 kinase. Use your left mouse button to rotate the molecule, zoom in by clicking on the left hand side of the display, scroll down the text on the left hand side and click on the blue links to see different annotated views of the protein, and right click on the graphical display for more option, and right click on the graphical display for more options.
Click here to view an example of our new ActiveICM product (US Patent No:7,880,738) in action. The files used in ActiveICM can be generated using ICM Browser Pro, ICM Chemist, or ICM Pro and ActiveICM is free to customers of this product. The online example is a Structural Genomics Consortium datapack file of PAK4 kinase. Use your left mouse button to rotate the molecule, zoom in by clicking on the left hand side of the display, scroll down the text on the left hand side and click on the blue links to see different annotated views of the protein, and right click on the graphical display for more option, and right click on the graphical display for more options.
| NOTE: The demo will only work if you download ActiveICM. Click here to download ActiveICM. |
 ICM now supports the 3D XML file format which is a popular 3D format for CAD. For example, Microsoft Virtual Earth uses this file format as well as CAD software companies such as Dassault Systems. There is a big online repository of 3DXML models here, models are free to download once you have registered. Once the file is downloaded, open the ICM graphical user interface, click on File/Open in ICM to bring the file into ICM and then display the file using the ICM Workspace.
ICM now supports the 3D XML file format which is a popular 3D format for CAD. For example, Microsoft Virtual Earth uses this file format as well as CAD software companies such as Dassault Systems. There is a big online repository of 3DXML models here, models are free to download once you have registered. Once the file is downloaded, open the ICM graphical user interface, click on File/Open in ICM to bring the file into ICM and then display the file using the ICM Workspace.
The Atomic Property Field (APF) superposition method is available in ICM Chemist Pro and the details of this method have been published this year by Dr. Maxim Totrov (Molsoft, Principal Scientist). Totrov M (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 71: 15-27
APF is a 3D pharmacophoric potential implemented on a grid. APF can be generated from one or multiple ligands and seven properties are assigned from empiric physico-chemical components (hydrogen bond donors, acceptors, Sp2 hybridization, lipophilicity, size, electropositive/negative and charge). Click here to see how to use the APF method.
 We now have over 80 members in the ICM Discussion Forum but this only represents a very small percentage of the ICM users worldwide. Click here to join the forum today. It is free to join the forum and there are many benefits of being part of this group: you will always be the first to get to know the new ICM features, you will learn new tips and tricks in ICM, find out how others are applying the software in their research, and most importantly you will be part of a network of people who can support and advise you when you have an ICM question. Click here to join the forum today.
We now have over 80 members in the ICM Discussion Forum but this only represents a very small percentage of the ICM users worldwide. Click here to join the forum today. It is free to join the forum and there are many benefits of being part of this group: you will always be the first to get to know the new ICM features, you will learn new tips and tricks in ICM, find out how others are applying the software in their research, and most importantly you will be part of a network of people who can support and advise you when you have an ICM question. Click here to join the forum today.
 Making Molecular Movies in ICM
Making Molecular Movies in ICM
To make a movie in ICM all you have to do is click on the movie button located at the bottom of the graphical user interface. Any animation or view in the ICM graphical window will then be dumped into a movie file. For more instructions please click here.
07.11.2008 July 2008 MolSoft ICM Newsletter
MolSoft ICM Newsletter July 2008
Welcome to the July 2008 MolSoft ICM Newsletter containing information on:
Fully Interactive 3D Ligand Editor
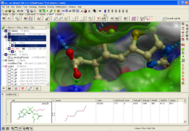 The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor. You can eidt the ligand by adding new functional groups, sampling a large collection of fragments on the fly, edit atoms or bonds, and and then in seconds see the affect of these changes on ligand binding. Clashes with the receptor are clearly flagged and energy contributions of each atom and total bidning energy and binding score are provided. Please see the graphical user interface manual for more information and instructions http://www.molsoft.com/gui/ligand-editor.html
The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor. You can eidt the ligand by adding new functional groups, sampling a large collection of fragments on the fly, edit atoms or bonds, and and then in seconds see the affect of these changes on ligand binding. Clashes with the receptor are clearly flagged and energy contributions of each atom and total bidning energy and binding score are provided. Please see the graphical user interface manual for more information and instructions http://www.molsoft.com/gui/ligand-editor.html
Docking a Markush Library
Generate a Markush library and dock a focused version of it on the fly. See this example, optimizing the roscovitine ligand bound to CDK5 http://www.molsoft.com/gui/markush-docking.html
Export Chemical Spreadsheets to Excel
Export the 2D chemical sketch as well as other column data in a spreadsheet directly to Microsoft Excel. See: http://www.molsoft.com/gui/export-excel.html
Read and Display Google Sketchup 3D Images
 You can now read, render and display any 3D object in ICM. e.g. KMZ and COLLADA format directly from the google sketchup website. We have also added options for changing the texture and material of a 3D object. See http://www.molsoft.com/gui/google-objects.html
You can now read, render and display any 3D object in ICM. e.g. KMZ and COLLADA format directly from the google sketchup website. We have also added options for changing the texture and material of a 3D object. See http://www.molsoft.com/gui/google-objects.html
Lots of New Data Plotting Features From ICM Tables
New plotting features include:
See http://www.molsoft.com/gui/table-plot.html for instructions
Background Images
 Add any image you want to the background. This is great for making cool images or you can directly compare molecules by making a background image and then making changes to the molecule and see clearly the differences. See http://www.molsoft.com/gui/background.html
Add any image you want to the background. This is great for making cool images or you can directly compare molecules by making a background image and then making changes to the molecule and see clearly the differences. See http://www.molsoft.com/gui/background.html
IUPAC Nomenclature
ICM will now determine the IUPAC nomenclature for any small molecule you sketch or read into ICM. See http://www.molsoft.com/gui/iupac.html
Ribbons and Chain Breaks
Display smooth ribbons and control the display of chain breaks See: http://www.molsoft.com/gui/structure-representation.html#ribbon
Local Chemical Databases
 In many cases a compound database (sdf file) is so large that you cannot open it in ICM because you do not have enough memory on your computer. A way around this was to place the database on our product called MolCart but now you can build local chemical databases which you can browse and edit.
In many cases a compound database (sdf file) is so large that you cannot open it in ICM because you do not have enough memory on your computer. A way around this was to place the database on our product called MolCart but now you can build local chemical databases which you can browse and edit.
Some of the features include:
For more information please click here http://www.molsoft.com/gui/local-databases.html
Occlusion Shading
 The occulusion shading option provides better representation of depth within a cavity. The color of each surface element of a grob (mesh) is changed by mixing its own color with the background depending on the burial of the surface element. See http://www.molsoft.com/gui/mesh-options.html#occlusion-shading
The occulusion shading option provides better representation of depth within a cavity. The color of each surface element of a grob (mesh) is changed by mixing its own color with the background depending on the burial of the surface element. See http://www.molsoft.com/gui/mesh-options.html#occlusion-shading
Protect Objects From Deletion
This option is useful when making molecular documents and you do not want the viewer of the document to delete an object that is contained in the document. See: http://www.molsoft.com/gui/make-molecular-document.html#protect
Wide-Screen Layout
There is now a new layout for users that have a widescreen. Try it by going to File/Preferences/Gui Tab and change GUI.windowLayout
New Command Line Options
Other new command line options are described in the release notes .
ActiveICM - Integrate ICM into PowerPoint or a Web Browser
 ActiveICM will change the way you give presentations or share ICM data with your colleagues. With ActiveICM you can display a series of slides generated in ICM inside PowerPoint or a web browser. The slides are fully interactive which means the user can manipulate the molecules in 3D inside the viewer.
Click here to wownload ActiveICM and see the instructions here http://www.molsoft.com/gui/activeicm.html
ActiveICM will change the way you give presentations or share ICM data with your colleagues. With ActiveICM you can display a series of slides generated in ICM inside PowerPoint or a web browser. The slides are fully interactive which means the user can manipulate the molecules in 3D inside the viewer.
Click here to wownload ActiveICM and see the instructions here http://www.molsoft.com/gui/activeicm.html
The next ICM workshop will be held in La Jolla CA on October 2nd to 3rd. The workshop will be presented by Prof. Ruben Abagyan (Scripps Research Institute, and Founder of MolSoft ) and Dr. Maxim Totrov (Principal Scientist, Molsoft). See http://www.molsoft.com/training.html
Quick Ligand and Peptide Pocket Display
 Did you know you can quickly display the surface of a ligand or peptide by pocket by right clicking on the ligand or peptide in the ICM workspace and selecting Ligand Pocket. See http://www.molsoft.com/gui/dsPocket.html
Did you know you can quickly display the surface of a ligand or peptide by pocket by right clicking on the ligand or peptide in the ICM workspace and selecting Ligand Pocket. See http://www.molsoft.com/gui/dsPocket.html
 | There were over 20,000 people at the Bio 2008 meeting and we thoroughly enjoyed demonstrating the latest ICM features and answering questions from the many people who stopped by at our booth. It was great to see so many ICM-users at the meeting and meet for the first time some of our international customers from Europe, Australia, and Japan. We received great feedback on our new ligand editor feature which will be released very soon, and our molecular document technology also attracted alot of interest. Other pictures from Bio2008 can be found here. |

05.22.2008 MolSoft will be Exhibiting at Bio2008
MolSoft LLC is looking forward to presenting at the Bio2008 conference in San Diego June 17-20. Please come and visit us at booth #2539 where we will be showcasing for the first time lots of exciting new tools that will be released later this year in ICM Version 3.5-2.
04.08.08 New ICM Paper: New method for the assessment of all drug-like pockets across a structural genome.
Nicola G, Smith CA, Abagyan R. New method for the assessment of all drug-like pockets across a structural genome. Comput Biol. 2008 Apr; 15(3):231-40.
Department of Molecular Biology, The Scripps Research Institute, La Jolla, California.
Abstract: With the increasing wealth of structural information available for human pathogens, it is now becoming possible to leverage that information to aid in rational selection of targets for inhibitor discovery. We present a methodology for assessing the drugability of all small-molecule binding pockets in a pathogen. Our approach incorporates accurate pocket identification, sequence conservation with a similar organism, sequence conservation with the host, and structure resolution. This novel method is applied to 21 structures from the malarial parasite Plasmodium falciparum. Based on our survey of the structural genome, we selected enoyl-acyl carrier protein reductase (ENR) as a promising candidate for virtual screening based inhibitor discovery.
03.20.08 New ICM Publication: A new method for ligand docking to flexible receptors
New ICM publication by the Scripps Research Institute and MolSoft scientists. See: Bottegoni G, Kufareva I, Totrov M, Abagyan R. A new method for ligand docking to flexible receptors by dual alanine scanning and refinement (SCARE). J Comput Aided Mol Des. 2008 Feb 14;
Paper link:
Abstract Protein binding sites undergo ligand specific conformational changes upon ligand binding. However, most docking protocols rely on a fixed conformation of the receptor, or on the prior knowledge of multiple conformations representing the variation of the pocket, or on a known bounding box for the ligand. Here we described a general induced fit docking protocol that requires only one initial pocket conformation and identifies most of the correct ligand positions as the lowest score. We expanded a previously used diverse "cross-docking" benchmark to thirty ligand-protein pairs extracted from different crystal structures. The algorithm systematically scans pairs of neighbouring side chains, replaces them by alanines, and docks the ligand to each 'gapped' version of the pocket. All docked positions are scored, refined with original side chains and flexible backbone and re-scored. In the optimal version of the protocol pairs of residues were replaced by alanines and only one best scoring conformation was selected from each 'gapped' pocket for refinement. The optimal SCARE (SCan Alanines and REfine) protocol identifies a near native conformation (under 2 A RMSD) as the lowest rank for 80% of pairs if the docking bounding box is defined by the predicted pocket envelope, and for as many as 90% of the pairs if the bounding box is derived from the known answer with approximately 5 A margin as used in most previous publications. The presented fully automated algorithm takes about 2 h per pose of a single processor time, requires only one pocket structure and no prior knowledge about the binding site location. Furthermore, the results for conformationally conserved pockets do not deteriorate due to substantial increase of the pocket variability.
03.11.08 Free Online Tutorial Dates Released for May through to July.
Click here for information on the upcoming online demonstrations.
03.03.08 New Review Article from MolSoft on Flexible Docking
Here is a link to a new publication in Current Opinion Structural Biology on flexible docking by Dr. Totrov (MolSoft) and Prof. Abagyan (Scripps). The paper highlights all the recent development for including multiple receptor conformations in docking to represent ligand induced fit.
02.14.08 New Tutorial: How to model the effect of a mutation on protein structure.
Click here for a new tutorial on how to make a mutation in a protein structure and optimize.
02.14.08 MolSoft Scientists Collaborate with Schering Plough to Identify New GPCR Inhibitors using ICM.
Click here to read how MolSoft and Schering Plough scientists identified new inhibtitors for a GPCR using MolSoft's drug discovery products. The structure-based development of MCH-R1 and other GPCR antagonists is hampered by the lack of an available experimentally determined atomic structure. A ligand-steered homology modeling approach has been developed (where information about existing ligands is used explicitly to shape and optimize the binding site) followed by docking-based virtual screening. Top scoring compounds identified virtually were tested experimentally in an MCH-R1 competitive binding assay, and six novel chemotypes as low micromolar affinity antagonist "hits" were identified. This success rate is more than a 10-fold improvement over random high-throughput screening, which supports our ligand-steered method. Clearly, the ligand-steered homology modeling method reduces the uncertainty of structure modeling for difficult targets like GPCRs.
02.04.08 New Tutorial: How to dock to multiple receptor conformations.
There is a new tutorial online here
The tutorial uses Aldose Reductase as an example. Aldose Reductase has a flexible loop in the ligand binding pocket vicinity which enables a variety of inhibitors to bind and therefore in order to identify these ligands via docking it is necessary to sample the conformations of this loop and dock to an ensemble of structures.
12.15.07 New Docking tutorials - Docking to Electron Density and Focused Markush Library Docking
How to dock a ligand to an electron density map. The ICM X-Ray AutoFit is an automated method to fit a ligand into electron density. The tool combines the powerful ICM docking algorithm with an electron density fitting function. Click here for step by step instructions.
How to dock a focused Markush library. A Markush library can be generated on the fly and docked using ICM. In this example we use the roscovitine ligand bound to CDK5 to identify scaffolds which may improve ligand-receptor interaction.
10.23.08 New Slide Transition EffectIn ICM version 3.5-1l a new blending transition effect between slides is available. To see this in action download the latest version of ICM or the free ICM-Browser and view this icb file:
http://www.molsoft.com/~andy/blend.icb
To generate this transition effect:
October 22nd 2007

As a quarter of million people fled their homes amid fierce wildfires that had burned 100,000 acres around San Diego County, Molsoft opened its doors for the families of the escapees. Children, pets, and adults have found a comfortable home at Molsoft and were given a place to stay, sleep, access internet and play video games.
Games, food, laughter and partial property destruction made their stay at Molsoft more comfortable. The training class became an information center from which the families were following the fate of their homes.
October 17th 2007
MolSoft launches the ICM Forum. Please sign up and contribute with questions/answers and interact with other ICM users worldwide.
October 12th 2007
MolSoft announces a series of online demonstrations as a new way of teaching the ICM suite of software. Click here for more information.
July 12th 2007
Molecular Movie Making Made Easy
Real-time screen grabbing movie making is now available in ICMBrowserPro on all platforms. With one click of the mouse anything displayed in the graphical displaywindow will be transfered to movie format - mpeg, avi or mov. See an example of an ICM movie here.
June 2nd 2007
MolCart Compound Database Update
The MolCart Compound screening library has been updated. The database contains 3,621,728 unique compounds for use in MolCart. Please contact info@molsoft.com for more information on licensing MolCart.
May 25th 2007
The latest version 3.5 for all ICM products has been released for Windows, Mac , Linux and SGI.Please download the latest version of ICM from our support center.
Some of the new features include:
For more details please see the Release Notes.
March 27th 2007 Maxim Totrov PhD to present data at American Chemical Society National Meeting
On March 27th 2007 Dr. Maxim Totrov (Principal Scientist - MolSoft LLC) will be giving a presentation entitled "Chemical superposition and pharmacophore elucidation by SCAPFOld: Self-consistent atomic property field optimization" at the American Chemical Society 233rd National Meeting & Exposition, Chicago, IL USA. The presentation abstract can be found here.
March 22nd to 23rd 2007 ICM Workshop
Once again the ICM Workshop sold out quickly and we had a full class of students eager to learn about Protein Structure and Drug Discovery using MolSoft's ICM software. The first day and a half of training was presented by Prof. Ruben Abagyan (Scripps Research Institute). Prof. Abagyan gave lectures and demonstrations on ICM sequence and structure analysis, molecular modeling and cheminformatics. Friday afternoon was devoted to small molecule docking, virtual ligand screening and protein-protein docking led by Dr. Maxim Totrov (Principal Scientist - MolSoft). The participants ran docking examples such as virtual screening of small ligand databases to identify known inhibitors of the cyclooxygenase receptor and protein-protein docking of subtilisin and chymotrypsin. All the participants agreed that the workshop will help them greatly when they return to the lab and would highly recommend the training to their colleagues.
If you were unable to attend this workshop we are holding workshops in May and September this year. Please click here for more details and sign up early to guarantee your place.
August 1st 2006 MolSoft Awarded NIH SBIR Phase I Grant to develop protein surface annotation software
MolSoft was recently awarded a NIH SBIR Phase I Grant to develop an integrated protein surface annotation suite of software for biologists and chemists. The software will allow users to apply several binding site prediction methods and analyze the results. These new algorithms include a protein-protein interface prediction method for small molecule binding sites and a surface structural motif detection and visualization method.
July 1st 2006 MolSoft Receives Bill and Melinda Gates Foundation AIDS Research Grant
Scientists at MolSoft LLC will be working as part of a multidisciplinary team led by Susan Zolla-Pazner Ph.D. (New York University School of Medicine) on a grant funded by the Bill and Melinda Gates Foundation entitled "The V3 Loop: A Conserved Structure of gp120 that Can Induce Broadly Neutralizing Antibodies Against HIV-1"
MolSoft LLC is excited to be leading the structure-based engineering of the V3-carrier immunogen. Using our experience with similar efforts we will undertake ab initio loop modeling of protein segments consisting of the central portions of the V3 signature sequences and modeling of carrier joining sequences. For more information please see the press release in the New York Times.
MolSoft and Virginia Tech will be working together on a 3-year project funded by the Foundation of the National Institutes of Healt to apply state-of-the-art computer modeling and chemical synthetic approaches to produce highly potent and selective anticholinesterases (AChEs) for malaria vector control. MolSoft's ICM software will be used to build homology models of AChEs and virtual screening will be applied to identify selective inhibitors. ICM-Chemistry will then be applied to optimize the lead compounds.
February 8th 2006: From Physics to Biology: the interface between experiment and computation BIFI 2006 - II International Conference, Zaragoza Spain.
Claudio Cavasotto PhD (Senior Scientist - Molsoft) has been invited to present a talk entitled "Ligand docking and virtual screening in structure-based drug discovery" at the BIFI 2006 - II International Conference, Zaragoza, Spain. His talk will be on February 8th 2006 and more details about the conference can be found here.
January 25th-27th 2006 Three-Day ICM Workshop.
 On January 25th-27th 2006, a 3-day ICM Workshop was held at Molsoft's La Jolla
offices. The first two days of the workshop covered all aspects of protein
structure and drug discovery including sequence analysis, protein modeling,
small molecule docking, protein-protein docking, cheminformatics and QSAR. The
third day was dedicated to advanced ICM training including the ICM command
language, scripting, loop modeling and flexible receptor docking. The course was conducted by Prof. Ruben Abagyan (The Scripps Research Institute) and
Dr. Maxim Totrov (Principal Scientist - Molsoft).
On January 25th-27th 2006, a 3-day ICM Workshop was held at Molsoft's La Jolla
offices. The first two days of the workshop covered all aspects of protein
structure and drug discovery including sequence analysis, protein modeling,
small molecule docking, protein-protein docking, cheminformatics and QSAR. The
third day was dedicated to advanced ICM training including the ICM command
language, scripting, loop modeling and flexible receptor docking. The course was conducted by Prof. Ruben Abagyan (The Scripps Research Institute) and
Dr. Maxim Totrov (Principal Scientist - Molsoft).
January 1st 2006 MolSoft and the Mayo Clinic College of Medicine Collaboration
Scientist at the Mayo Clinic and MolSoft will be collaborating on a project to model G-protein coupled receptors (GPCRs) which are key receptors for a large number of disease pathways. The aim is to use MolSoft's proprietary software to build highly-refined and rigorously validated models of GPCRs and to use these models to guide experimental work at the Mayo Clinic.
September 29th-30th 2005 Successful Workshop
On September 29th-30th an international group of ICM users from academia and industry gathered for our ICM Workshop entitled "Protein Structure and Drug Discovery" Click here to see what went on.
Molsoft has released the latest version (3.4) of ICM. This version includes a number of new features as described in the Release Notes. The most significant addition is the incorporation of ICM Molecular Documents and Presentation (US Patent No:7,880,738). These can be constructed and viewed in ICM, they can consist of text linked to molecular animations and transitions. The animations and transitions are fully-interactive and can be interrupted without any loss of information. For more information on ICM Molecular Documents and Presenations please click here.
August 10th, 2004
Press Release:San Diego, CA. Molsoft LLC and the Structural Genomics Consortium at the University of Oxford Announce Software Co-Development and Research Collaboration. more details
Molsoft is pleased to learn that the Department of Chemistry and Biochemistry at California State University, Fullerton, is expanding its teaching use of ICM-Pro. CSU Fullerton began using ICM-Pro for training undergraduate chemistry and biochemistry students in Spring 2004. ICM-Pro has also been used to teach structural bioinformatics to students seeking a Certificate in Bioinformatics through CSUFs Extended Education program. Beginning in 2005-06, ICM-Pro will be featured in a new computational biology program for chemistry and biochemistry majors [more].
On the 20-21st of July 2004 sixteen people attended the ICM workshop entitled "In Silico Drug Discovery". The workshop was held at Bristol Myers Squibb at Princeton New Jersey.
Molsoft LLC is pleased to announce a collaboration with the Burnham Institute, La Jolla, on a STTR NIH funded grant entitled "15 - Deoxy -12,14-prostaglandin J2 as a ligand of RXRalpha".
March 17, 2004 Molsoft LLC Receives Phase I STTR to Develop Beta-Catenin Antagonists
Molsoft LLC received a Phase I STTR award from the National Institutes of Health, National Cancer Institute, for the project entitled. Rational Development of TCF/Beta-Catenin Antagonists. The project will combine Molsoft's novel in silico lead development platform with the cancer biology resources at The Burnham Institute to discover and characterize small molecule ligands using the beta-catenin structure both alone and in complex with TCF. Beta-catenin signaling has been implicated in a number of malignancies, which makes the beta-catenin/TCF interaction an promising target for the prevention and treatment of a variety of tumors and leukemia, and the project proposes to develop a novel generation of drugs active against various forms of cancer.
July 25, 2003 Molsoft LLC Receives Small Business Biodefense Grant Award
Molsoft LLC received a Small Business Biodefense Program award from the National Institutes of Health, National Institute of Allergy and Infectious Diseases, for the project entitled .Rational Design of Inhibitors of Yersinia pestis EF-Tu.. Molsoft and its research partners at the University of California, Irvine and Chemical Diversity Labs, Inc., San Diego, propose to develop a new class of antibacterial agents active against Category A pathogens likely to be used by bioterrorists including those causing plague, anthrax, cholera, and typhoid fever. Furthermore, the project proposes to develop design principles that will be useful in developing agents less susceptible to the problem of rapidly emerging antibiotic drug resistance.
Mpex Pharmaceuticals Inc. (ìMpexî) and Molsoft, LLC (ìMolsoftí) have announced a collaboration to rationally design new antibacterial agents based on the refinement of a 3-D molecular structure of a membrane protein target, the functional characterization of its surfaces, and the application of predictive algorithms ..more..
June 16, 2003 Molsoft LLC Receives Phase II STTR Grant Award to Continue Developing Thyroid Receptor Antagonists
Molsoft LLC received a Phase II STTR award from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, for continuation of the "Rational Development of Thyroid Receptor Antagonists. project. In Phase I, Molsoft and its collaborators at the NYU School of Medicine and the NYU Department of Chemistry achieved proof of concept. Molsoft.s virtual ligand screening technology was used to discover 14 small molecule thyroid receptor antagonists displaying extreme structural diversity with IC50s ranging from 4 to 30 microM. Additionally, the test-case lead optimization scheme designed in Phase I based on generating focused virtual libraries of molecules easily amenable to organic parallel synthesis resulted in the rapid identification of second generation hits with IC50 in the nanomolar range. In Phase II, Molsoft and its NYU research partners propose to conduct full-scale optimization cycles of selected hits identified in Phase 1 in order to produce low nanomolar hits and to evaluate those hits using computational tools, in vitro characterization, and preliminary animal studies.
21 October 2002. Molsoft to Evaluate Novel Scaffolds for PDF Inhibitor Program
Molsoft LLC, a La Jolla based company providing new, breakthrough technologies in computational chemistry and biology, and GeneSoft Pharmaceuticals Inc., a specialty pharmaceutical company headquartered in South San Francisco, announced that they have signed a collaborative agreement to evaluate inhibitors of peptide deformylase (PDF), an essential bacterial metalloenzyme and novel antibiotic target ..more...
Molsoft conducted an intensive 2-day training course/workshop on the ICM suite of products at La Jolla, CA. The lecture and hands-on demonstrations covered topics of interest to computational chemists and biologists in the area of drug discovery. The topics included molecular graphics, animation, bioinformatics, homology, protein modeling, docking and ICM command language for expert users.
The attendance was international with scientists coming from Asia and Europe as well as from various parts of the US. The attendees were interested in having future courses, both at the basic and at the advanced levels.
One of the attendees from Australia who is very well known in the modeling field for his early work in structural bioinformatics, Dr. Jiri Novotny (Consultant for Biocomputing and Bioinformatics), said, "The course was well organized and presented the participants with two full days of new and efficient tools applicable to the most pressing problems of current structural biology and bioinformatics: structural/functional annotation of genomic data, analysis of protein surfaces, ligand binding sites, virtual ligand screening, structure derivation, regularization and model building". He continued on to say, "Perhaps the most impressive aspect of the workshop was the skill, dedication and the highest professional standard of the scientific team established at Molsoft by Prof. Abagyan. In hands-on session at computer screens the participants were encouraged to ask questions and bring out problems that were answered and solved immediately by the whole team including themselves. It was an invaluable practice and a good demonstration of the speed and versatility of the newest ICM version."
 The new version, ICM 3.0, was introduced at the course and was well accepted as
an extremely user-friendly interface by the attendees. This version, which also
has stereo viewing mode that works with VRex glasses on Linux and Windows
platforms, was welcomed by all the participants as an efficient and low cost
alternative to existing stereo viewing tools.
The new version, ICM 3.0, was introduced at the course and was well accepted as
an extremely user-friendly interface by the attendees. This version, which also
has stereo viewing mode that works with VRex glasses on Linux and Windows
platforms, was welcomed by all the participants as an efficient and low cost
alternative to existing stereo viewing tools.

About the training course, Dr. Maxim Totrov, co-founder and Principal Scientist at Molsoft said, "The course provided us with a vital opportunity to meet members of the growing community of ICM-users and bring them up-to date with the latest developments in ICM. It was exciting to see first-time users perform complex molecular modeling tasks such as protein-ligand docking after only a few hours of training. These workshops are geared towards helping our customers utilize fully the power of ICM molecular modeling suite in their everyday work."
22 August 2002. Molsoft Participates in the Annual Symposium of the Protein Society in San Diego
 Molsoft is pleased to announce its participation at the Protein Society's 16th
Annual Symposium held at the Marriott in San Diego from August 17-21, 2002.
Attendance was estimated at around 1300. Molsoft's exhibit was designed to
inform Protein Society attendees about the latest technology available from
Molsoft including its recent version of a new graphical interface which enables
Molsoft's ICM software easier to use than ever before.
Molsoft is pleased to announce its participation at the Protein Society's 16th
Annual Symposium held at the Marriott in San Diego from August 17-21, 2002.
Attendance was estimated at around 1300. Molsoft's exhibit was designed to
inform Protein Society attendees about the latest technology available from
Molsoft including its recent version of a new graphical interface which enables
Molsoft's ICM software easier to use than ever before.
"We enjoyed talking to numerous visitors to our booth," said Dr. Lalitha Subramanian, Director of Contract Research for Molsoft, "many of whom were extremely excited to see ICM's new GUI interface." She added, "Our science has always been strong and well validated. Now, with the added power of a very user friendly interface, ICM will be the choice for most researchers in the academic and commercial fields."
Molsoft also participated in the Beyond Genome 2002 conference held in June of this year and will be presenting at the BioITWorld conference to be held in November.
Molsoft received a Phase II SBIR award from the Department of Energy for continuation of the "GAP: Genomics Annotation Platform" project. In Phase I Molsoft used its many years of experience in developing molecular visualization and manipulation software to built the computational tools central to the functionality of the GAP. In Phase I, Molsoft successfully used GAP to predict the function of uncharacterized genes. In Phase II, Molsoft will optimize the existing tools and develop an efficient graphical user interface while adding a system for automatic updates to GAP. When completed, the Molsoft GAP will provide an online research platform allowing users from the pharmaceutical industry to access Molsoft's state-of-the-art computational biology technology and to use that technology for extracting meaningful information from the human genome project and from other major, international sequencing efforts.
Molsoft received a Phase 1 SBIR grant from the National Institutes of Health, National Human Genome Research Institute for its project entitled "Sequence/Structure Annotation of Protein Families." The goal in this project is analyzing in silico well known protein families in order to identify which family members could be good therapeutic targets and how pharmacogenomics strategies can be devised to better design drugs targets at these proteins.
August 2001
Molsoft and Plexus Vaccine Inc.entered into a licensing agreement to discover and commercialize new interventional strategies for global infectious diseases, for drug-resistant microbes, and for chronic diseases associated with cryptic pathogens. Plexus will use the Molsoft ICM software suite to develop Virtual Epitope databases of molecular disease targets for in silico design of structural mimics that can be readily synthesized, tested, and formulated as vaccines ..more..
August 2001
Molsoft received a Phase 1 SBIR grant from the Department of Energy for its project entitled "GAP: Genomics Annotation Platform." The goal in this project is to develop bioinformatics tools used for predicting the function of uncharacterized genes.
Molsoft and HTS Biosystems, Inc. (HTS) entered into collaboration for Molsoft to develop software modules for instrument control, data collection, local visualization, and analysis of data generated by HTS' Surface Plasmon Resonance (SPR) instruments.
April 2001
Molsoft LLC and Chemical Diversity Labs, Inc. (CDL), San Diego, CA, formed a strategic alliance to provide joint chemistry and computational modeling services to their customers. Molsoft will use known three-dimensional structures of drug targets as the starting point for the rational selection process. The Molsoft modeling by homology technology will be applied in cases when the structure is not available. CDL will give Molsoft an access to its collection of over 250,000 diverse purified small molecules already available for biological screening.
February 2001
Molsoft and Biovitrum AB, a subsidiary of Pharmacia & Upjohn AB, entered into an agreement to co-develop a Chem-Informatics Client System. Molsoft and Biovitrum will use Molsoft's proprietary programming languages and libraries to develop the software, which will be called BeeHive and will function as a program manager and database interface for chemical searches, analysis, and integration.
November 2000
Molsoft has moved to a new location, click the press release for more details.
October 2000
Molsoft and Syrrx, Inc., a pioneer in the field of structural proteomics, entered into a strategic alliance to accelerate structure-guided drug discovery through the combination of Molsoft's Virtual Ligand Screening (VLS) technology and other computational biology technologies with Syrrx's high-throughput structural proteomics platform. ..more..
September 2000
Molsoft received a Phase 1 STTR grant from National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases for its project entitled "Rational Development of Thyroid Receptor Antagonists." The goal of this project is to discover and optimize small molecule antagonists of the thyroid hormone receptor. Molsoft will work with the New York University School of Medicine to complete the project.
September 2000
Molsoft LLC announced the relocation of its headquarters and commercial development operations to an expanded facility in La Jolla. The new facility will accommodate several expanded programs, including the continued refinement of ICM 2.8, the expansion of software sales and the addition of new products and in-house services. The new facilities include 3500 square feet of computer laboratory, research, training, and office space in close proximity to The Scripps Research Institute and the Genomics Institute of the Novartis Research Foundation (GNF), both of whom have research collaboration agreements with Molsoft.
September 2000
Molsoft LLC announced its first shipment of the Molsoft BioPackage to Hitachi, Ltd. for distribution in Japan. During the two-year Distribution Agreement, which became effective June 1, 2000, Hitachi will be the exclusive distributor of Molsoft's BioPackage in Japan. Hitachi plans to market the software to Japanese corporations, universities, and laboratories. Molsoft's computer software provides novel computational and informational technologies for biomedicine including genomics and drug discovery. ..more..
April 2000
Molsoft and The Genomics Institute of the Novartis Research Foundation (GNF) have entered into a Research Collaboration Agreement. The collaboration will focus on the further development of computational biology tools for functional and structural annotation of new genomic sequences, protein modeling, and virtual ligand screening. Molsoft will provide its ICM software for research in functional and structural annotation of new genomic sequences, protein modeling and lead discovery at GNF, which will provide GNF researchers with an accurate, speedy and flexible method for structural annotation and protein structure modeling. Molsoft will grant GNF a nonexclusive license to use Molsoft databases and software in gene discovery, functional genomics, and drug discovery, and GNF will supply support for personnel and computer facilities to be used in the joint tool development projects ..more..
July 2000
Molsoft LLC and The Scripps Research Institute (TSRI), a private, non-profit research organization engaged in basic biomedical research, announced the formation of research collaboration. During the term of the agreement, TSRI and Molsoft will collaborate on defined joint projects involving the functional and structural annotation of new genomic sequences and protein modeling. For the joint projects, Molsoft will grant TSRI a nonexclusive and restricted license to use Molsoft databases and software in gene discovery, functional genomics and drug discovery, and TSRI will supply personnel and computer facilities for use in the joint projects. ..more..
November 1999
Molsoft LLC and eBioinformatics, Inc. announced the formation of an alliance in which Molsoft will provide its ICM software for incorporation into eBioinformatics, Inc. flagship product BioNavigator. The Molsoft-ICM suite of computational biology tools will provide BioNavigator users with access to the latest and most powerful algorithms. This alliance will give scientists new opportunities based on the power of Molsoft's ICM software and the convenience of BioNavigator's web-based bioinformatics workspace ..more..