ICM-VLS (Virtual Ligand Screening) |
NOTE: This product is an add-on to ICM-Pro. This product also includes all the chemistry functionalities included in ICM-Chemist and ICM-Chemist-Pro.
ICM Virtual Ligand Screening (VLS) is a combination of the genuine internal coordinate docking methodology with a sophisticated global optimization scheme. Accurate and fast potentials and empirically adjusted scoring functions have led to an efficient virtual screening methodology in which ligands are fully and continuously flexible. The average yield of lead candidates is as high as 10% and in many cases only the top 1% of hits need to be tested experimentally to find a lead. The product includes:
Features
Results Analysis Features
In 2017 and 2018, MolSoft ranked in first place in both the D3R Grand Challenge 2 and 3 for docking pose prediction and affinity prediction. The papers describing the results and methods were published:
In 2018 - MolSoft ranked in first place in an independent evaluation of six covalent docking methods. ICM outperformed the other programs in all tests including reproducing the experimental binding modes and scoring. In an interesting side test ICM performed considerably better than GLIDE for non-covalent conventional docking.
Novartis Scientists Identify 3 Lead Compounds using ICM-VLS in World's Largest VLS Screen At the recent Amazon Cloud Computing conference ( AWS Re:Invent ) scientists at Novartis reported using MolSoft's ICM-VLS in the largest ever virtual screen. MolSoft's software developers built a cloud-version of ICM-VLS which enabled Novartis to undertake this ground breaking achievement in cloud-based virtual screening. The jobs were distributed to more than 30,000 cores in the USA and Europe. Novartis undertook 10.6 years of drug compound computations in 9 hours! The virtual screen resulted in three new lead compounds which are now being optimized by Novartis. This work was previously reported at MolSoft's user group meeting earlier this year and at BioIT 2012. Watch video of presentation - click on image (right).
At the recent Amazon Cloud Computing conference ( AWS Re:Invent ) scientists at Novartis reported using MolSoft's ICM-VLS in the largest ever virtual screen. MolSoft's software developers built a cloud-version of ICM-VLS which enabled Novartis to undertake this ground breaking achievement in cloud-based virtual screening. The jobs were distributed to more than 30,000 cores in the USA and Europe. Novartis undertook 10.6 years of drug compound computations in 9 hours! The virtual screen resulted in three new lead compounds which are now being optimized by Novartis. This work was previously reported at MolSoft's user group meeting earlier this year and at BioIT 2012. Watch video of presentation - click on image (right).
|
|
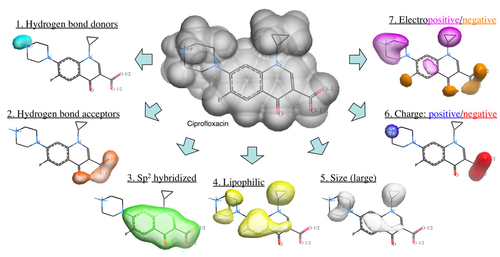
Ligand-based virtual screening can be performed using the Atomic Property Field (APF) method developed by MolSoft ( Totrov 2008). APF is a 3D pharmacophoric potential implemented on a continuously distributed grid which can be used for ligand docking and scoring. APF can be generated from one or more high affinity scaffolds and seven properties are assigned from empiric physico-chemical components. These properties include: hydrogen bond donors, acceptors, Sp2 hybridization, lipophilicity, size, electropositive/negative and charge (image right). A single ligand atom can contribute to multiple fields; multiple similar ligand atoms in a spatially consistent location result in a strong pharmacophore signal for their features in this location. APF has also been extended to multiple flexible ligand alignments using an iterative procedure. APF uses Monte Carlo minimization in the atomic property fields potentials in conjunction with standard force-field energies. [ More...] |
| Title | Authors | Journal | Year | PubMed | Notes |
|---|---|---|---|---|---|
| Discovery of a Natural Product-Like iNOS Inhibitor by Molecular Docking with Potential Neuroprotective Effects In Vivo | Zhong HJ, Liu LJ, Chong CM, Lu L, Wang M, Chan DS, Chan PW, Lee SM, Ma DL, Leung CH. | PLoS One. 2014 | 2014 | 24690920 | |
| Structure-based discovery of a small non-peptidic Neuropilins antagonist exerting in vitro and in vivo anti-tumor activity on breast cancer model. | Borriello L, Montes M, Lepelletier Y, Leforban B, Liu WQ, Demange L, Delhomme B, Pavoni S, Jarray R, Boucher JL, Dufour S, Hermine O, Garbay C, Hadj-Slimane R, Raynaud F. | Cancer Lett. 2014 | 2014 | 24752068 | |
| Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. | Bavan S, Sherman B, Luetje CW, Abaffy T. | PLoS One. 2014 | 2014 | 24637889 | |
| Identification and Characterization of Small Molecules That Inhibit Nonsense-Mediated RNA Decay and Suppress Nonsense p53 Mutations. | Martin L, Grigoryan A, Wang D, Wang J, Breda L, Rivella S, Cardozo T, Gardner LB. | Cancer Res. 2014 | 2014 | 24662918 | |
| Identification of novel serotonin transporter compounds by virtual screening. | Gabrielsen M, Kurczab R, Siwek A, Wolak M, Ravna AW, Kristiansen K, Kufareva I, Abagyan R, Nowak G, Chilmonczyk Z, Sylte I, Bojarski AJ. | J Chem Inf Model. 2014 | 2014 | 24521202 | |
| High-throughput virtual screening identifies novel N'-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible LSD1 inhibitors. | Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, Sharma S. | J Med Chem. 2013 | 2013 | 24237195 | |
| Structure-based ligand discovery targeting orthosteric and allosteric pockets of dopamine receptors. | Lane JR, Chubukov P, Liu W, Canals M, Cherezov V, Abagyan R, Stevens RC, Katritch V. | Mol Pharmacol. 2013 | 2013 | 24021214 | |
| Identification of a new class of FtsZ inhibitors by structure-based design and in vitro screening. | Chan FY, Sun N, Neves MA, Lam PC, Chung WH, Wong LK, Chow HY, Ma DL, Chan PH, Leung YC, Chan TH, Abagyan R, Wong KY. | J Chem Inf Model. 2013 | 2013 | 23848971 | |
| Specific small molecule inhibitors of Skp2-mediated p27 degradation. | Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. | Chem Biol. 2012 | 2012 | 23261596 | |
| Small molecule inhibitors of Bacillus anthracis protective antigen proteolytic activation and oligomerization. | Wein AN, Williams BN, Liu S, Ermolinsky B, Provenzano D, Abagyan R, Orry A, Leppla SH, Peredelchuk M. | J Med Chem. 2012 | 2012 | 22954387 | |
| A metal-based inhibitor of tumor necrosis factor-?Ą. | Leung CH, Zhong HJ, Yang H, Cheng Z, Chan DS, Ma VP, Abagyan R, Wong CY, Ma DL. | Angew Chem Int Ed Engl. 2012 | 2012 | 22807261 | |
| Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. | Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. | Nat Chem Biol. 2011 | 2011 | 21706033 | |
| In search of allosteric modulators of a7-nAChR by solvent density guided virtual screening. | Dey R, Chen L. | J Biomol Struct Dyn. 2011 | 2011 | 21294583 | |
| A natural product-like inhibitor of NEDD8-activating enzyme. | Leung CH, Chan DS, Yang H, Abagyan R, Lee SM, Zhu GY, Fong WF, Ma DL. | Chem Commun (Camb). 2011 | 2011 | 21240405 | |
| The pthaladyns: GTP competitive inhibitors of dynamin I and II GTPase derived from virtual screening. | Odell LR, Howan D, Gordon CP, Robertson MJ, Chau N, Mariana A, Whiting AE, Abagyan R, Daniel JA, Gorgani NN, Robinson PJ, McCluskey A. | J Med Chem. 2010 | 2010 | 20575553 | |
| Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. | Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, Cole PA. | Chem Biol. 2010 | 2010 | 20534345 | Screened 500K compounds and selected 194 for experimental testing resulting in 3 inhibitors which had specificity. |
| Structure-based discovery of natural-product-like TNF-?Ą inhibitors. | Chan DS, Lee HM, Yang F, Che CM, Wong CC, Abagyan R, Leung CH, Ma DL. | Angew Chem Int Ed Engl. 2010 | 2010 | 20235259 | |
| Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. | Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R. | J Med Chem. 2010 | 2010 | 20095623 | |
| Structural determinants of PERK inhibitor potency and selectivity. | Wang H, Blais J, Ron D, Cardozo T. | Chem Biol Drug Des. 2010 | 2010 | 21070610 | Specific inhibitors identified by ICM-VLS. |
| Identification of new classes of ricin toxin inhibitors by virtual screening. | Bai Y, Watt B, Wahome PG, Mantis NJ, Robertus JD. | Toxicon. 2010 | 2010 | 20493201 | |
| Chromene-3-carboxamide derivatives discovered from virtual screening as potent inhibitors of the tumour maker, AKR1B10. | Endo S, Matsunaga T, Kuwata K, Zhao HT, El-Kabbani O, Kitade Y, Hara A. | Bioorg Med Chem. 2010 | 2010 | 20304656 | Discovery of several chomene-3carboxamide derivatives as potent competitive inhibitors. |
| Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. | Bisson WH, Koch DC, O'Donnell EF, Khalil SM, Kerkvliet NI, Tanguay RL, Abagyan R, Kolluri SK. | J Med Chem. 2009 | 2009 | 19719119 | Discovery of new class of AhR inhibitors. |
| A novel small-molecule inhibitor of the avian influenza H5N1 virus determined through computational screening against the neuraminidase. | An J, Lee DC, Law AH, Yang CL, Poon LL, Lau AS, Jones SJ. | J Med Chem. 2009 | 2009 | 19419201 | Used MolSoft?s ICMPocketFinder to identify a new pocket conformation. Used ICM-VLS to identify a ligand with different binding pose and interactions than oseltamivir and zanamivir. |
| In silico screening for PTPN22 inhibitors: active hits from an inactive phosphatase conformation. | Wu S, Bottini M, Rickert RC, Mustelin T, Tautz L. | ChemMedChem. 2009 | 2009 | 19177473 | Sub and low micormolar inhibitors discovered using ICM-VLS. |
| Discovery of potent thermolysin inhibitors using structure based virtual screening and binding assays. | Khan MT, Fuskev?Łg OM, Sylte I. | J Med Chem. 2009 | 2009 | 19072688 | NCI compound library screened and 12 inibitors discovered. |
| Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. | Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, O'Neill KA, Hine H, Burton MS, Voigt JH, Abagyan RA, Bayne ML, Monsma FJ Jr. | J Med Chem. 2008 | 2008 | 18198821 | First demonstration that GPCR models can be used for antagonist discovery by virtual screening. |
| Discovery of small molecule inhibitors of ubiquitin-like poxvirus proteinase I7L using homology modeling and covalent docking approaches. | Katritch V, Byrd CM, Tseitin V, Dai D, Raush E, Totrov M, Abagyan R, Jordan R, Hruby DE. | J Comput Aided Mol Des. 2007 | 2007 | 17960327 | 230,000 available ketone and aldehyde compounds were screened. Out of 456 predicted ligands, 97 inhibitors of I7L proteinase activity were confirmed in biochemical assays. |
| De novo discovery of serotonin N-acetyltransferase inhibitors. | Szewczuk LM, Saldanha SA, Ganguly S, Bowers EM, Javoroncov M, Karanam B, Culhane JC, Holbert MA, Klein DC, Abagyan R, Cole PA. | J Med Chem. 2007 | 2007 | 17924613 | 1.2 million compounds were screened and 241 compounds tested resulting in the discovery of a new class of inhibitors. |
| Small molecules block the polymerization of Z alpha1-antitrypsin and increase the clearance of intracellular aggregates. | Mallya M, Phillips RL, Saldanha SA, Gooptu B, Brown SC, Termine DJ, Shirvani AM, Wu Y, Sifers RN, Abagyan R, Lomas DA. | J Med Chem. 2007 | 2007 | 17918823 | ICM-VLS was performed on 1.2 million small molecules and 6 antagonists were identified which were further optimized using ICM tools. |
| Discovery of antiandrogen activity of nonsteroidal scaffolds of marketed drugs. | Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, May LT, Christopoulos A, Dalton JT, Sexton PM, Zhang XK, Abagyan R. | Proc Natl Acad Sci U S A. 2007 | 2007 | 17606915 | Screening to multiple receptor conformations of the androgen receptor led to the identification of an antagonist. |
| Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. | Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. | Science. 2007 | 2007 | 17588900 | |
| Discovery of novel inhibitors targeting enoyl-acyl carrier protein reductase in Plasmodium falciparum by structure-based virtual screening. | Nicola G, Smith CA, Lucumi E, Kuo MR, Karagyozov L, Fidock DA, Sacchettini JC, Abagyan R. | Biochem Biophys Res Commun. 2007 | 2007 | 17509532 | ChemBridge database was screened. 169 compounds were tested experimentally and 16 compounds had activity. |
| In silico prediction of SARS protease inhibitors by virtual high throughput screening. | Plewczynski D, Hoffmann M, von Grotthuss M, Ginalski K, Rychewski L. | Chem Biol Drug Des. 2007 | 2007 | 17461975 | |
| Discovery of bioactive small-molecule inhibitor of poly adp-ribose polymerase: implications for energy-deficient cells. | Altmann SM, Muryshev A, Fossale E, Maxwell MM, Norflus FN, Fox J, Hersch SM, Young AB, MacDonald ME, Abagyan R, Kazantsev AG. | Chem Biol. 2006 | 2006 | 16873024 | |
| In silico identification of novel EGFR inhibitors with antiproliferative activity against cancer cells. | Cavasotto CN, Ortiz MA, Abagyan RA, Piedrafita FJ. | Bioorg Med Chem Lett. 2006 | 2006 | 16413185 | 300K compounds were screened > 7 micromolar hits identified. |
| Small molecule inhibitors of dynamin I GTPase activity: development of dimeric tyrphostins. | Hill T, Odell LR, Edwards JK, Graham ME, McGeachie AB, Rusak J, Quan A, Abagyan R, Scott JL, Robinson PJ, McCluskey A. | J Med Chem. 2005 | 2005 | 16302817 | |
| Nuclear hormone receptor targeted virtual screening. | Schapira M, Abagyan R, Totrov M. | J Med Chem. 2003 | 2003 | 12825943 | |
| Discovery of diverse thyroid hormone receptor antagonists by high-throughput docking. | Schapira M, Raaka BM, Das S, Fan L, Totrov M, Zhou Z, Wilson SR, Abagyan R, Samuels HH. | Proc Natl Acad Sci U S A. 2003 | 2003 | 12777627 | 250K compounds were screened, 75 were tested experimentally and 14 antagonists were discovered. |
| In silico discovery of novel retinoic acid receptor agonist structures. | Schapira M, Raaka BM, Samuels HH, Abagyan R. | BMC Struct Biol. 2001 | 2001 | 11405897 | |
| Identification of ligands for RNA targets via structure-based virtual screening: HIV-1 TAR. | Filikov AV, Mohan V, Vickers TA, Griffey RH, Cook PD, Abagyan RA, James TL. | J Comput Aided Mol Des. 2000 | 2000 | 10921774 | High enrichment factor and 7 new inhibitors identified. |
| Rational discovery of novel nuclear hormone receptor antagonists. | Schapira M, Raaka BM, Samuels HH, Abagyan R. | Proc Natl Acad Sci U S A. 2000 | 2000 | 10655475 |
"I had a tough case of tightly bound ligands in a crystal structure whose bound poses were difficult to reproduce with other programs. They could just not find the right pose - often, ICM fit it right into this tight site, with terrific overlap with much of the crystal structure pose.
Dr. Terry R. Stouch, PhD, Head, Computational Chemistry, Lexicon Pharmaceuticals
| NOTE: Installation instructions for a linux cluster can be found here. |