MolScreen Contains a Panel of >2500 High Quality 2D and 3D Models.
MolScreen is a set of high quality 2D fingerprint and 3D pharmacophore models for a broad range of pharmacology and toxicology targets.
The models can be used for lead discovery or counter screening. The models use MolSoft's 2D QSAR/Fingerprint and 3D Atomic Property Fields ( Totrov 2008) methods. There are currently approximately 2500 models for 1200 targets.
The models can be screened directly using MolSoft's ICM-Pro + VLS software. Alternatively we can screen a set of chemicals for you via our contract research services. Please contact us for more information about how to use MolScreen.
MolScreen Applications
MolScreen can be used for:
- Target Identification - Search chemicals against a set of protein targets.
- Lead Identification - Identify chemicals that can bind against a protein target.
- Profiling - Multiple protein targets versus multiple chemicals.
- Drug-Repurposing - Use MolScreen to search for new protein targets for available drugs
Available Models
You can download and view the available models using the links below (updated (3/8/2019). Each model has a name which starts with the 3 letter abbreviation of the model type as described below followed by the gene name.
Model Types
There are two categories of models:
1. ADMET Prediction Models ( mcp)
- CACO2, hERG, HALFLIFE, LD50, CYP, Tox21, etc...
- Regression and Classification Models as well as fully connected neural networks.
2. Different types of Activity Models for a large Panel of Drug Targets
- Approximately 2500 models against 1200 targets.
- Machine Learning ( kcc), Ligand Field - 3D Atomic Properties Field ( dfz), 4D Docking/3D-QSAR ( dpc), 3D APF/3D-QSAR ( dfa) and Neural Network Chemical Classification ( ncc )
About the Models
Machine Learning Models - Hybrid 2D QSAR/Fingerprint Models kcc(+kca)
|
kcc(+kca): Kernel regression Chemical fingerprint Classification/Activity prediction
- Currently: 999 mammalian models
- Training set: ChEMBL Ki, IC50, EC50
- Report kcc(Classification) score and kca(pKd regression) score
- Median training set: 370 ligands
- Median external test set AUC: 96%
- Median external test set Q2: 0.5
- Extremely fast (thousands of cpds in min)
Training:
- Cluster Actives by fingerprint
- Add 40k ChEMBL actives decoy
- Kernel function to each cluster -> probability score (kcc/MolClass Score)
- Partial Least Square Regression for each cluster + Kernel Regression (kca/MolpKd Score)
- MolScore: combine MolpKd and MolSimilarity to known binders
|
Ligand Field Docking Models - 3D Atomic Property Field Models (dfz)

|
dfz: Docking to ligand Field Z-score prediction model
- Built using Atomic Property Fields
- Currently: 504 mammalian models
- Pocketome ligands/custom alignment as APF template
- ChEMBL cpds for validation
- Median AUC: 92%, 139 cpds vs decoy
- Fast-ish (single template cluster ~5 sec per cpd)
|
Pocket Docking 3D QSAR Models
|
dpc: Docking to Pocket Classification/Activity
- Currently: 343 mammalian models w/ AUC> 80%
- Training set: ChEMBL Ki, IC50, EC50, Drugbank assignment
- Median size: 307 ligands
- Median external Q2: 0.53
- Median external AUC: 95%
Training:
- Pocketome -> Clustering of pocket residues
- 4D Docking w/ co-crystallized ligand as APF template
- Docking Score -> Probability score (dpc/MolClass score)
- 3D QSAR training of Activity-> (dpa/MolpKd)
- MolScore: combine MolpKd and MolSimilarity to known binders
|
Hybrid 4D/2D - Hybrid Models (dfa)
|
dfa: Docking to ligand Field Activity prediction
- Currently: 612 mammalian models w/ AUC > 80%
- Training set: ChEMBL Ki, IC50, EC50, Drugbank assignment
- Median size: 270 ligands
- Median external Q2: 0.65
- Median external AUC: 96%
Training:
- Also from Pocketome -> 4D Docking + Ligand APF template
- Cpd align to ligand template -> cluster by 3D poses
- APF Score -> Probability Score (dfc/MolClass score)
- 3D-QSAR training for each cpd cluster (dfa/MolpKd score)
- MolScore: combine MolpKd and MolSimilarity to known binders
|
Neural Network - 2D Fingerprint Neural Network Classifier (ncc)
|
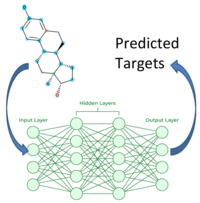
ncc: Neural Network Chemical fingerprint Classification.
- Currently: 6 Target Families each with 12-234 targets and 3K to 144K ligands.
- All Models are validated with 25% set aside as external test set.
- Median external AUC: 99.5%
Training for each family:
- Data: Targets(m) x Compounds(n)
- Input Layer: ECFP
- Fully Connected Neural Net with 2-3 hidden layers
- Output Layer: m Targets
- Multitask Prediction
|
ADMET Models
mcp: Miscellaneous Chemical Property Models
- Currently 38 models, mostly from PubChem data
- All validated by external test set (20% of data set aside)
- Regression Models, Mean external test set Q2: 0.7 - CACO2, PAMPA permeability, LD50 (mg/kg), Half-life (hr)
- Classification Models, Median external test set AUC: 84% - hERG, PGPinhibitor, PGPsubstrate, PAINS - Cytochrome P450 1A2, 2C19, 2C9, 2D6, 3A4 - 25 Tox21 Classifier, including Estrogen Agonist/Antagonist, Genotoxicity, Aromatase, etc
- Fully connected Neural Network models.

