Google Search: Keyword Search:
| Prev | ICM User's Guide 21.4 Sequence and Alignment Tutorials | Next |
[ Load and Superimpose PDB Structures | Link Sequence to Structure | Pocket Sequence Conservation | Alignment Annotation ]
The key topics in this tutorial are:
- How to extract sequences from PDB structures.
- How to load sequences from UniProt.
- How to make an alignment.
- How to link the alignment to 3D structure.
- How to display sequence conservation in ligand binding pocket.
- How to color 3D structure by alignment.
21.4.1 Display and Superimpose Protein Kinase Structures in Preparation for the Tutorial |
In preparation for the tutorial we will read in two PDB file and superimpose them. We will then extract sequences from the PDB structures and read in additional kinase sequences from Uniprot.
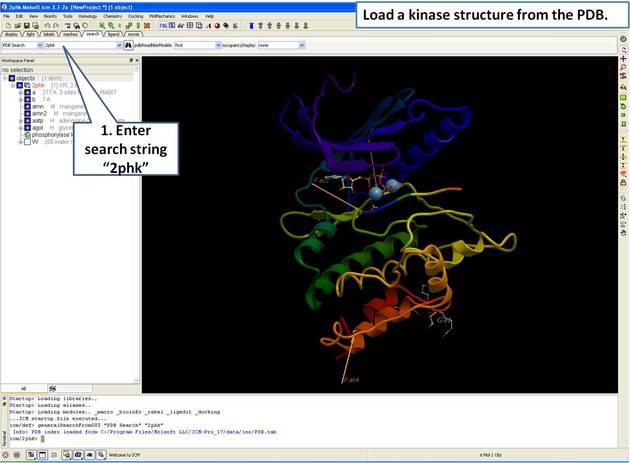 |
| Step 1: Load and display the kinase structure 2PHK. |
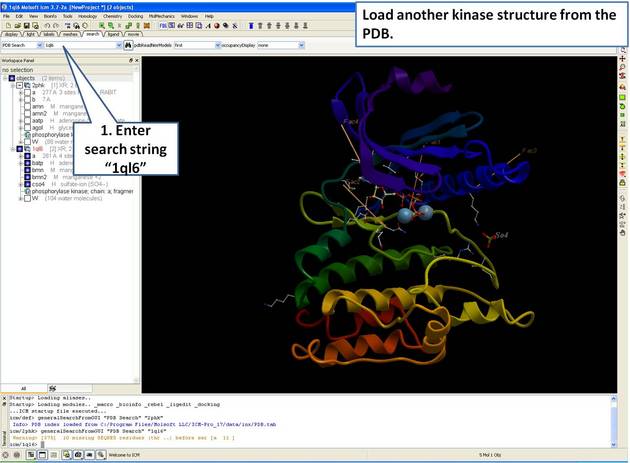 |
| Step 2: Load and display the kinase structure 1ql6. |
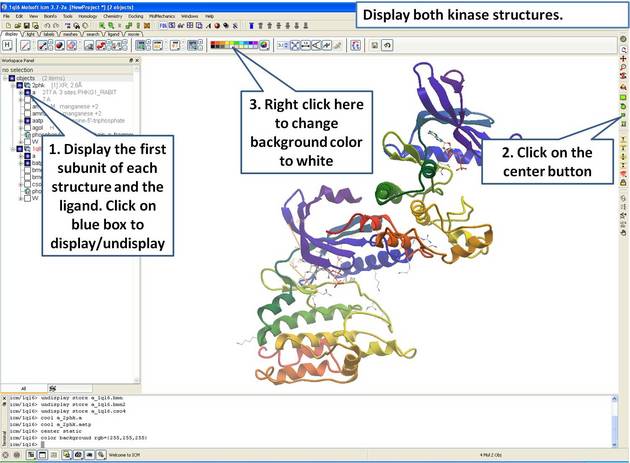 |
| Step 3: Display both receptors. |
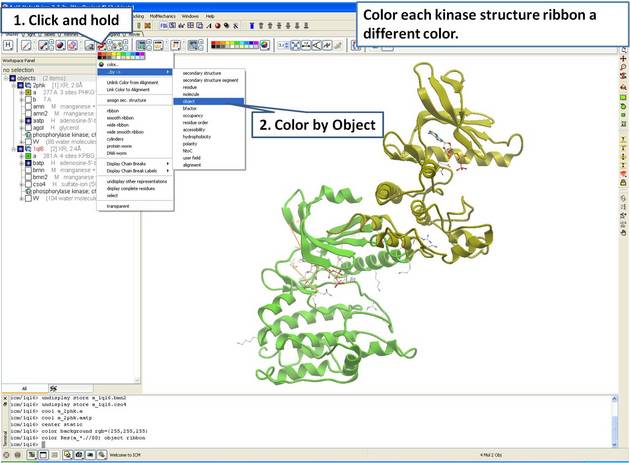 |
| Step 4: For clarity, color each receptor a different color. |
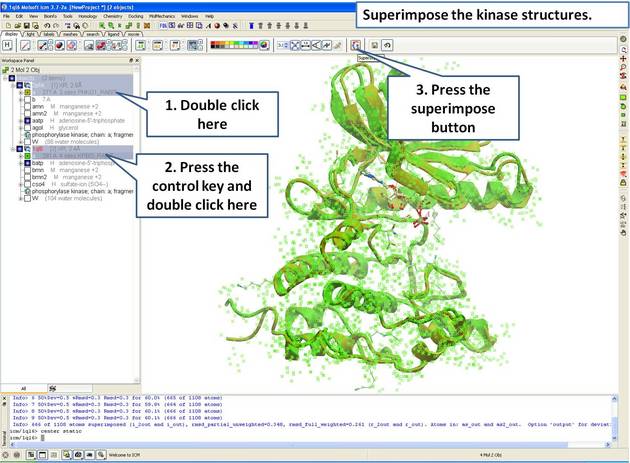 |
| Step 5: Superimpose both kinase structures using the superimpose button in the display tab. |
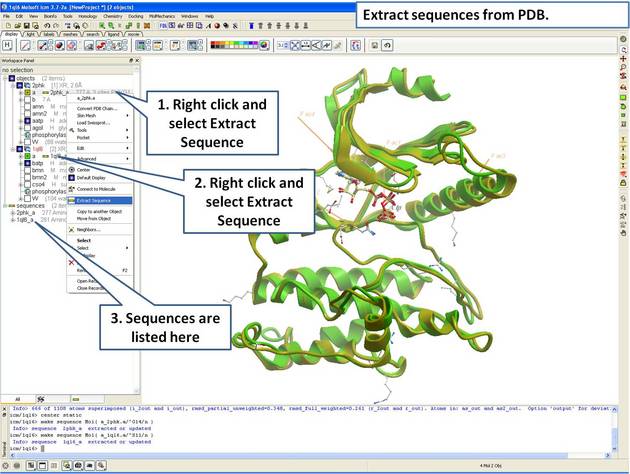 |
| Step 6: Extract the sequences from the PDB structure. |
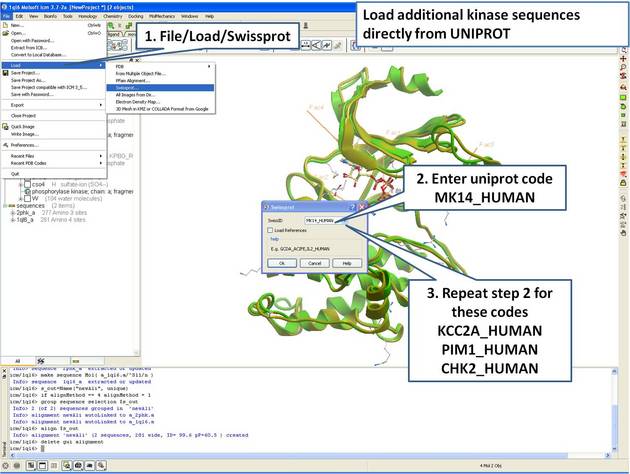 |
| Step 7: Load UniProt sequences. |
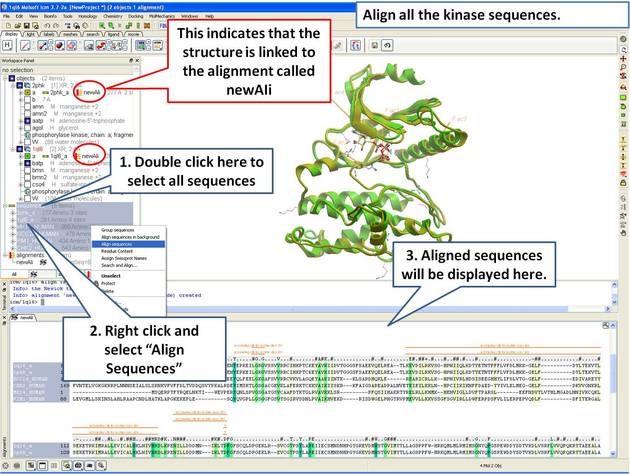 |
| Step 8: Make a sequence alignment. |
21.4.2 Linking Sequence Alignment to Structure |
An alignment can be directly linked to the 3D structure. If the sequence has been extracted from a displayed PDB file this will be done automatically. If multiple alignments are loaded you can change the link using Bioinfo/Link to Structure... menu. This examples continues from the previous section.
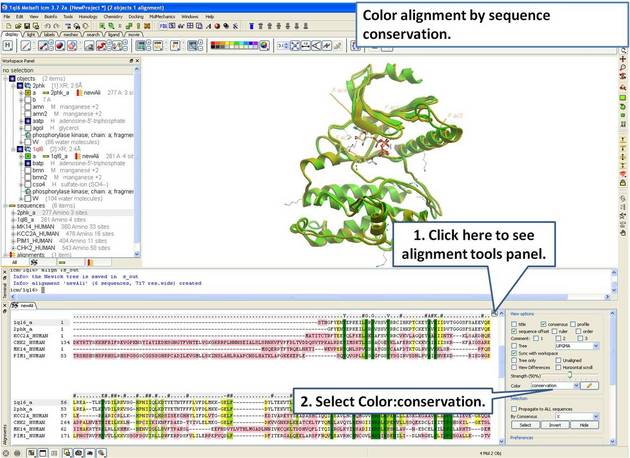 |
| Step 9: Color the alignment by sequence conservation. |
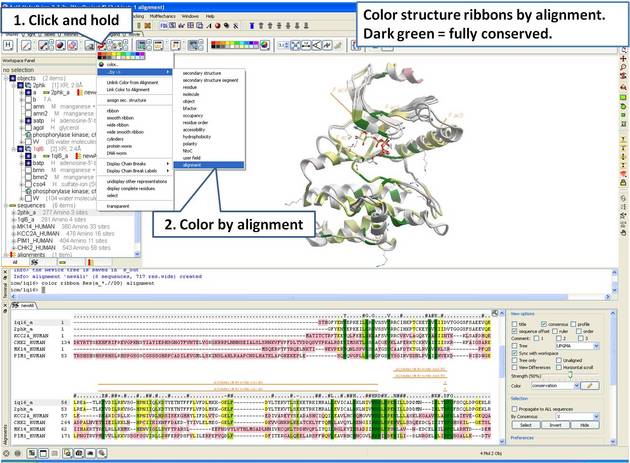 |
| Step 10: Color the 3D structure by the alignment color. |
21.4.3 Identify Sequence Conservation in Ligand Binding Pocket |
In this example we show how to display sequence conservation in the ligand binding pocket. This examples continues from the previous section.
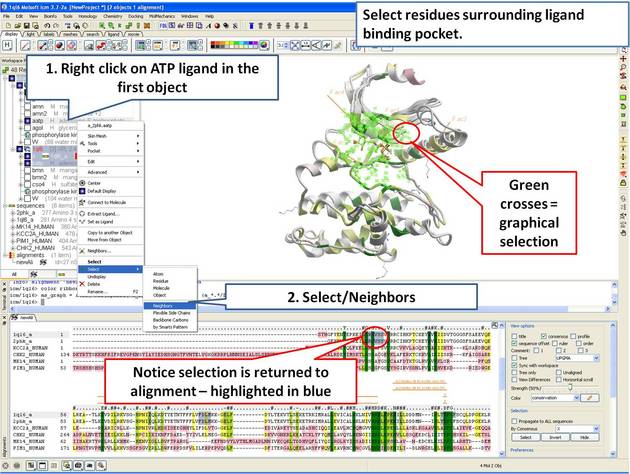 |
| Step 11: Select the residues surrounding the ligand binding pocket (or any other selection of interes). |
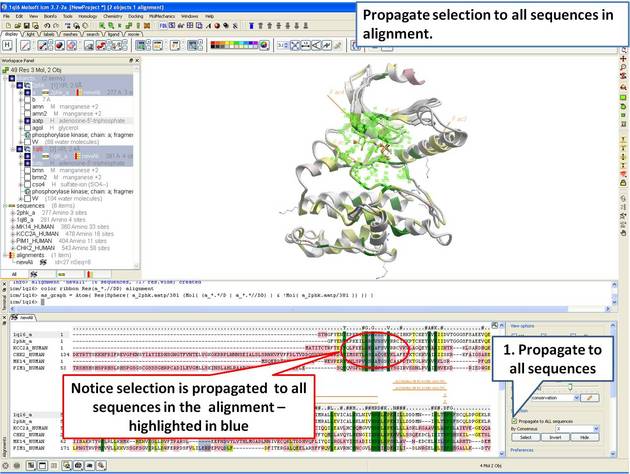 |
| Step 12: The 3D structure and alignment are linked, therefore the selections made in 3D are transferred to the alignment. Only the sequences that have 3D structures will be selected in the alignment, therefore we need to propagate the selection to all sequences in the alignment. |
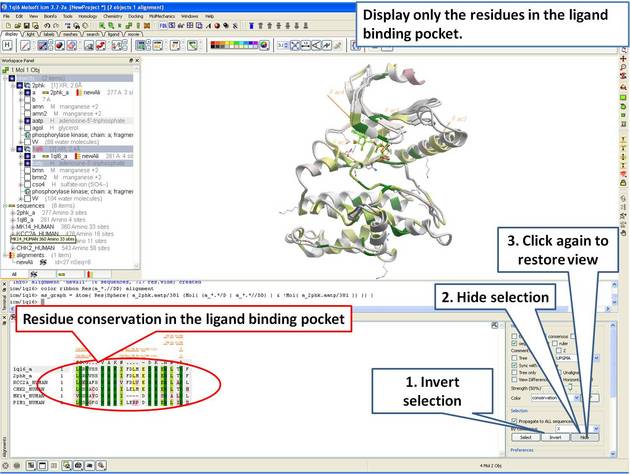 |
| Step 13: Display only the residues in the pocket in the alignment. |
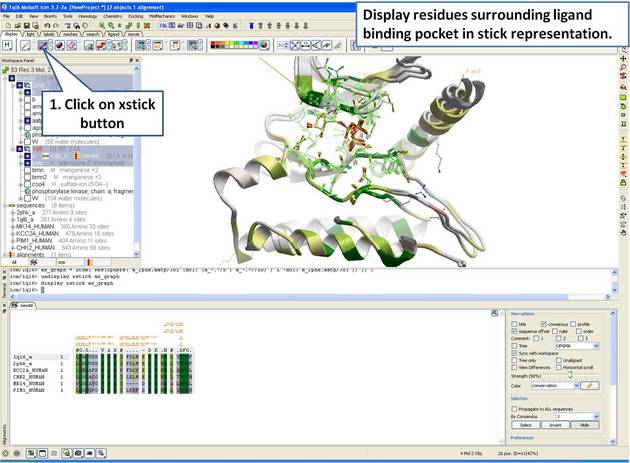 |
| Step 14: Display the residues surrounding the pocket in stick representation. |
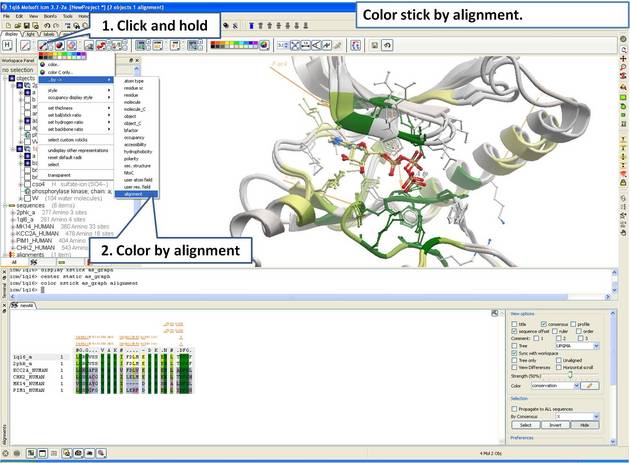 |
| Step 15: Color stick by the alignment color. |
21.4.4 Alignment Annotation |
Introduction
G-Protein Coupled Receptors (GPCRs) all share a common structural core of seven transmembrane helices but they lack significant sequence homology between subfamilies. When modeling GPCRs it is important to get a good alignmnent between the query and template structure. Each helix has one or more conserved motifs:Helix 1: GX3N or GN
Helix 2: N(S,H)LX3DX7,8,9P
Helix 3: SX3LX2IX2D(E,H)RY
Helix 4: WX8,9P
Helix 5: FX2PX7Y
Helix 6: FX2CW(Y,F)XP
Helix 7/Helix 8: LX3NX3N(D)PX2YX5,6F
The ProSite class A alignment http://prosite.expasy.org/PDOC00210 can be used to guide GPCR alignments. For this example we have prepared an icb file containing a subset of the class A GPCR sequences from ProSite. We will use this file to annotate the key conserved motifs this will help with the GPCR modeliing example. The file can be downloaded here: https://molsoft.com/addons/ftp/workshop (1. cut and paste the address into a web browser. 2. Right click on gpcr_ClassA and choose "Save Link As". 3. Open the .icb in ICM File/Open).
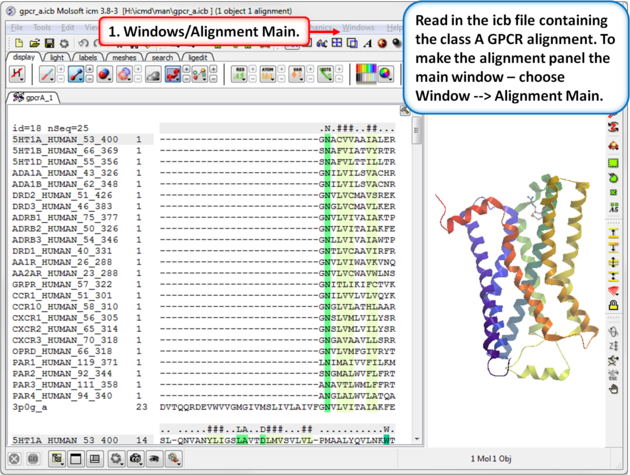 |
| Step 1: Read in the class A GPCR alignment (gpcr_ClassA.icb - see link above). Make the alignment the main window (Windows/Alignment Main). |
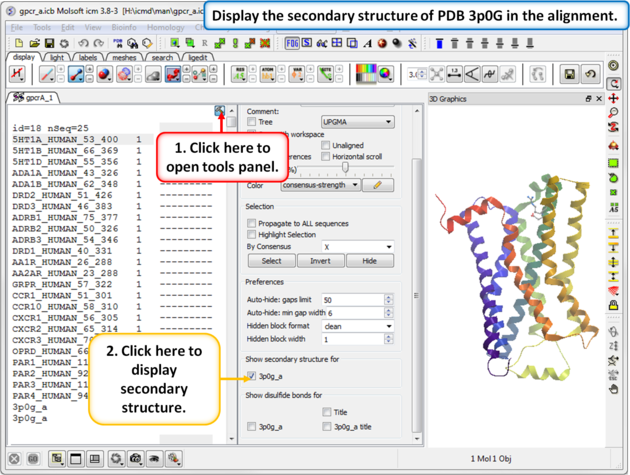 |
| Step 2: The sequence of the beta 2 adrenoceptor is appended to the alignment. This allows us to display the secondary structure of the protein in the alignment. |
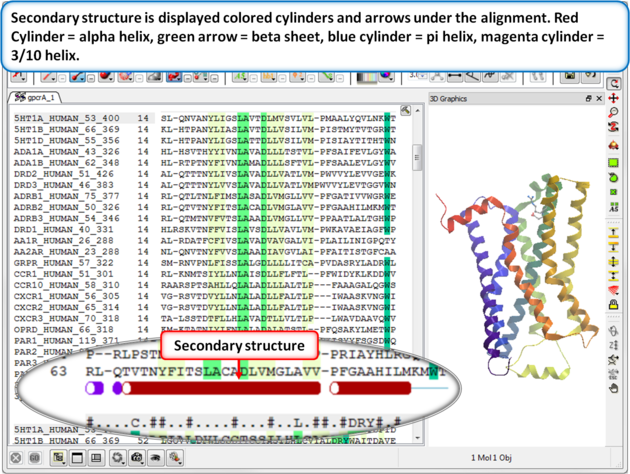 |
| Step 3: Secondary structure is displayed as colored cylinders and arrows under the alignment. Red Cylinder = alpha helix, green arrow = beta sheet, blue cylinder = pi helix, magenta cylinder = 3/10 helix. |
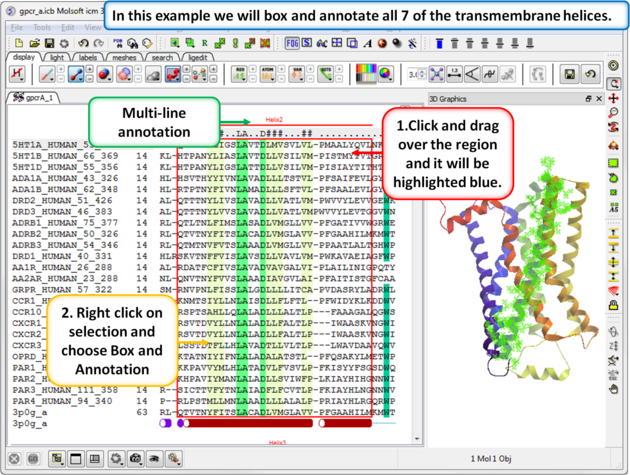 |
| Step 4: Box and annotate the seven transmembrane helices. |
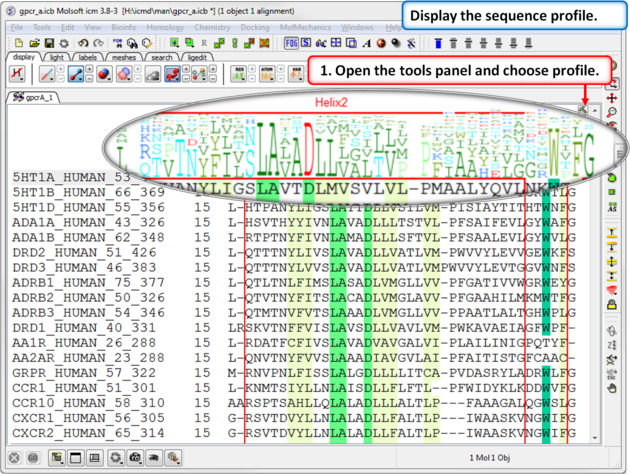 |
| Step 5: A sequence profile helps to get a better understanding of the sequence conservation at each point. |
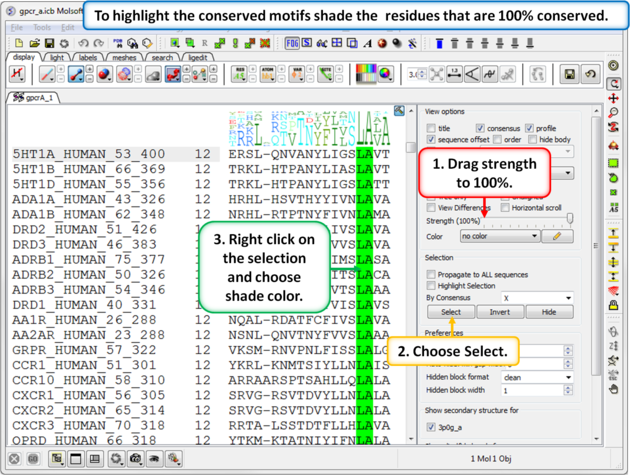 |
| Step 6: To highlight the conserved sequence motifs - set the consensus strength to 100% and then color the fully conserved residues. You may need to lower the conservation strength to pick out all the motifs. |
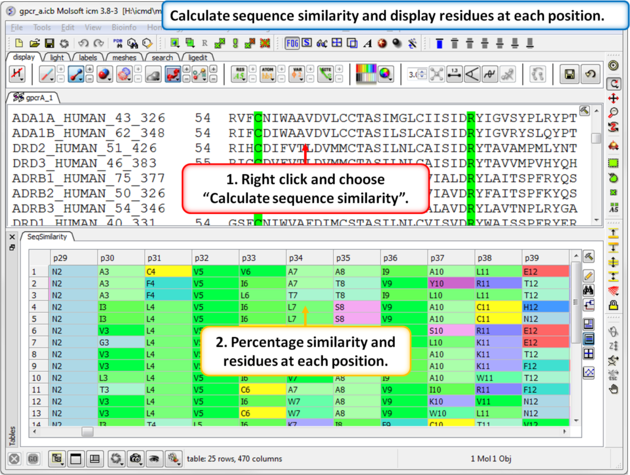 |
| Step 7: Calculate the sequence similarity and display the alignment residue profile. |
| Prev Biomolecule | Home Up | Next Homology Modeling Tutorials |