Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 5.7 3D Similarity PDB Search | Next |
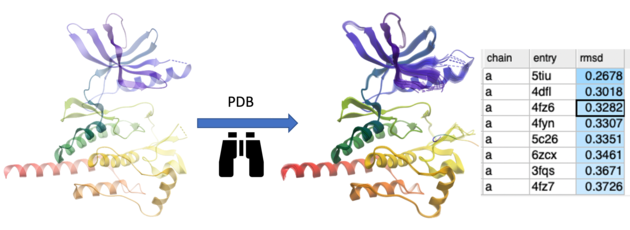
This option searches the PDB for proteins with similar structures or folds to the query. You can search using the entire protein, a specific region, or a non-contiguous selection (e.g., a ligand-binding pocket). The method first performs a fast 3D similarity search on a backbone-stripped version of the PDB, using distance RMSD between C-alpha atom distance matrices. For the top matches, the optimal coordinate RMSD is then calculated and reported.
- First read in your query protein structure.
- You can use selection to define the region of interest - whether it’s the whole protein, a specific segment, or a non-contiguous selection such as a ligand-binding pocket. election (e.g. ligand binding pocket).
- You can then right click and choose "3D protein similarity search" or select it in the Tools menu.
- A dialog box will be displayed and there is an option to update PDB index. First update your index - it might take a few minutes.
- Then select the rmsd search threshold - ICM will not report any hits higher than the threshold
- Click OK to run the search.
- A table will then be displayed reporting the hits ranked by rmsd.
- Double click to load the structure and superimpose it onto your query protein.
| Prev Identify Ligand Pockets | Home Up | Next Scan Pockets |