ICM Success Stories |
Documented here are a number of computational biology and chemistry success stories using MolSoft's ICM products. Please click on the image to read more and see the related references. Other peer-reviewed publications using ICM products are listed here.
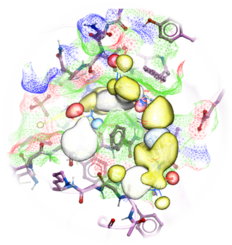 | MolSoft Outperforms a Range of other Methods for Docking Pose and Affinity Prediction Accuracy in D3R Grand Challenge 3 Scientists at MolSoft led by Max Totrov Ph.D. participated in the Drug Design Data Resource (D3R) Grand Challenge 3 (GC3) and once again achieved great success in the accuracy of docking pose prediction and ligand binding affinity. The competition is a "blinded" open challenge for the worldwide computational chemistry community bringing together teams using different docking methods and software. . Read more... |
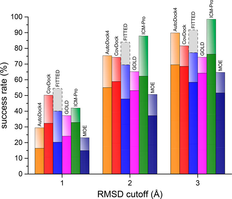 | MolSoft ICM ranks First Place in Covalent Docking Method Comparison A recent publication by Scarpino et al compared the performance of six covalent docking methods. ICM ranked first place in accuracy in all tests reported. Read more... |
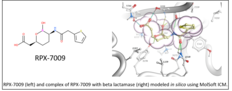 | FDA approves Vabomere: structure-based design using Molsoft's ICM was key to its discovery. Vabomere is a combination of meropenem (previously developed carbapenem antibiotic) and vaborbactam (RPX-7009), a novel cyclic boronate beta-lactamase inhibitor. Structure-based design by MolSoft's scientists (lead by Maxim Totrov Ph.D.) using the ICM-Pro desktop modeling software was key to the discovery of RPX-7009. The novel cyclic boronate chemotype was first modeled in-silico at Molsoft and prioritized for synthesis based on modeling and docking into several beta-lactamase target enzymes.. Read more... |
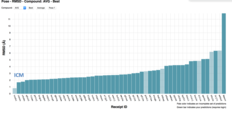 | MolSoft ICM D3R Docking Challenge Success Along with over 50 other participants, the Molsoft group led by Maxim Totrov Ph.D.(Principal Scientist, MolSoft), submitted blind docking pose predictions for the Farnesoid X receptor (FXR) which is a drug target for dyslipidemia and diabetes. The MolSoft team used the pocketome (www.pocketome.org) entry for FXR and the ICM-VLS, Atomic Property Fields (APF) and machine learning methods in the ICM-Pro software to predict the interaction of 36 FXR ligands and the binding affinity of 102 other ligands. Read more... |
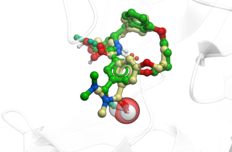 | Macrocycle Modeling - MolSoft ICM-Dock Again Performs Well in Industry-Wide Grand Challenge Competition The results of the "blinded" D3R Grand Challenge 4 ligand docking competition have been published and MolSoft once again has outperformed a range of other methods for ligand binding pose and activity prediction. Read more... |
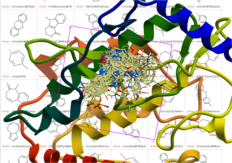 | Virtual Ligand Screening Success Stories MolSoft's ICM-VLS software is successfully applied worldwide by academic laboratories and the pharmaceutical industry for the identification of new lead compounds for drug design. There are many published examples where ICM-VLS has identified new compounds for a variety of drug targets including challenging ones such as GPCRs, nuclear receptors and kinases. Read more... |
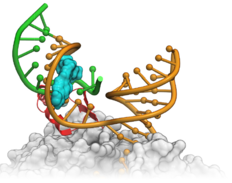 | RNA Modeling and Lead Optimization. Targeting RNA by small molecules is a high risk endeavour but potentially very rewarding. You can read an interesting story about RNA drug hunters here featuring some nice ICM graphics. Here we outline a couple of success stories by Novartis and Nymirum who used MolSoft's ICM for RNA docking, modeling and screening. Read more... |
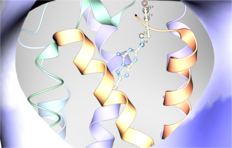 | MolSoft ICM Software Performs Very Well in Latest GPCR Modeling and Docking Competition. Two teams from Molsoft using ICM-Pro + VLS competed in the "blind" GPCR modeling competition to predict the structure and ligand interactions of the adenosine receptor. MolSoft's teams built models that had the largest number of correct ligand-receptor interatomic contacts, 45 and 34 out of 70, respecitvely out of all participants. Moreover, both models were ranked number 1 in the set of submitted complexes. Read more... |
 | Family A GPCR Modeling and Lead Compound Discovery Successes Scientists at MolSoft were the first to use an homology model of a GPCR to identify new lead compounds. ICM-Pro has also been used to better understand the agonist conformation of a GPCR. Read more... |
 | Family B GPCR Modeling Family B GPCRs are a challenging target for drug design due to a lack of atomic resolution structures. But by using extensive experimental data including cross-linking, FRET distances, mutagenesis, and structure activity relationship data useful validated models have been constructed. Read more... |
 | Kinase Modeling and Drug Discovery A number of methods have been developed using ICM-Pro to account for the flexibility of the ATP binding pocket and for the identification of new more specific binding sites in kinases. Read more... |
 | Nuclear Receptor Modeling and Virtual Screening ICM-Pro has a long history of success stories in nuclear receptor ligand binding prediction and modeling. Including modeling the antagonist form of the estrogen receptor and discovery of novel retinoic acid receptor antagonists by virtual ligand screening. Read more |
 | Development of Successful Methods for Induced-Fit Docking A number of successful methods have been developed by MolSoft to incorporate receptor flexibility (induced fit) upon ligand binding. These methods include incorporating multiple receptor conformations or using a known ligand to mold the receptor binding pocket. Read more... |
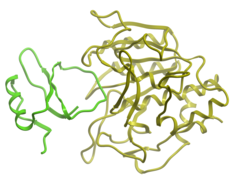 | Protein-Protein Docking Achievements The ICM-Pro software package has a long history of successes in predicting protein-protein complexes. In 1994 the software correctly predicted an antibody-lysozyme complex and more recently performed very well in worldwide protein-protein docking competitions. Read more... |
 | Breakthroughs in Molecular Graphics and Visualization MolSoft has always been a leader in breakthrough molecular graphics display. In the last few years the technology has also been used to communicate live fully-interactive 3D molecules via the web and PowerPoint. Read more on how you can do this here. |