Google Search: Keyword Search:
| Prev | ICM User's Guide 12.8 Covalent Docking | Next |
[ Sketch Reaction ]
| Available in the following product(s): ICM-Pro | ICM-VLS | |
To undertake covalent docking you should:
- Setup the docking project (Docking/SetProject).
- Setup the receptor (Docking/Setup Receptor) - you can remove the check mark in the "Make Receptor Maps Immediately" option because for covalent docking the maps are made at a later stage.
- Select the residue that will be modified (makes covalent bond) in the docking project receptor (e.g. mydock_rec).
- Choose the menu option Docking/Covalent and a dialog window as shown below will be displayed.
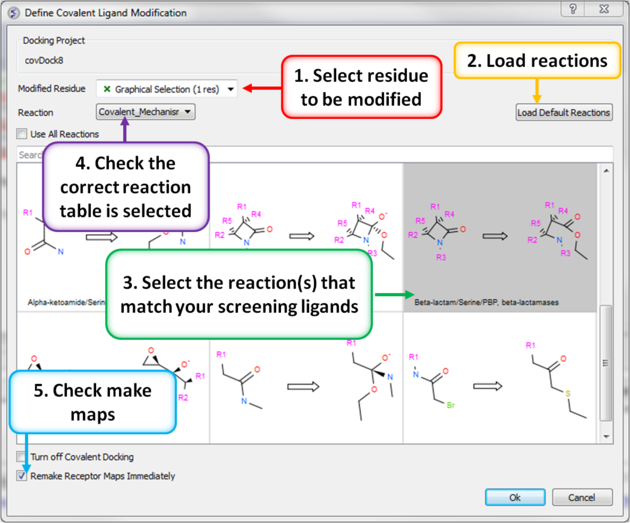 | |
|
- Next read in an SDF file containing the chemicals you wish to dock that match the reactant of the reaction.
- To dock the ligands choose Docking/Dock from Table.
To turn off covalent docking:
- Docking/Covalent docking and check the option "Turn off Covalent Docking".
12.8.1 Sketch a Reaction |
| NOTE: Save time! Please check the in-built reactions in ICM to check if your reaction has already been drawn. |
In this example we are sketching the general reaction equation for acylation of a serine side-chain by beta-lactam:
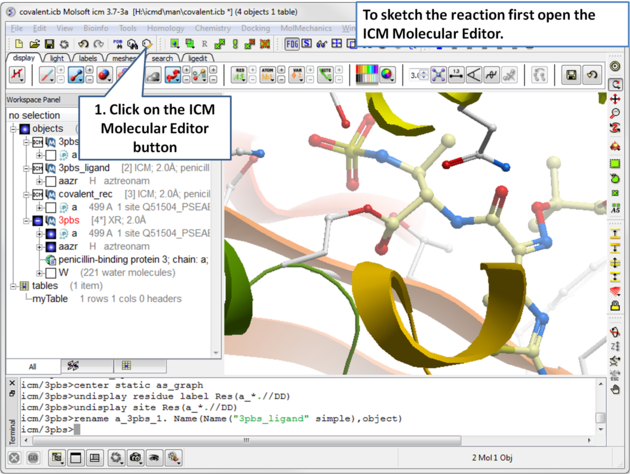 |
| Open the ICM Molecular Editor. |
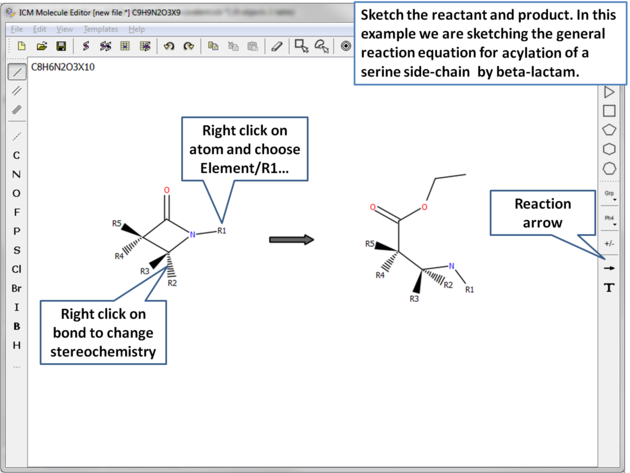 |
| Sketch the reactant and product. |
| Note: The reaction table may contain multiple reaction mechanisms, in which case the system will try to apply them sequentially to the ligand, until first matching reaction is found. This is useful if ligands with different types of 'warheads' (reactive groups) will be considered. |
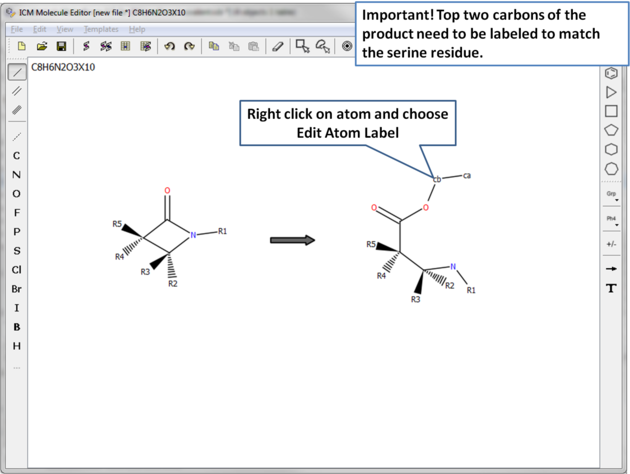 |
| Important! The top two atoms in the product which correspond to the modified serine residue need to be labeled to match the residue. In this example the atoms are labeled ca and cb. |
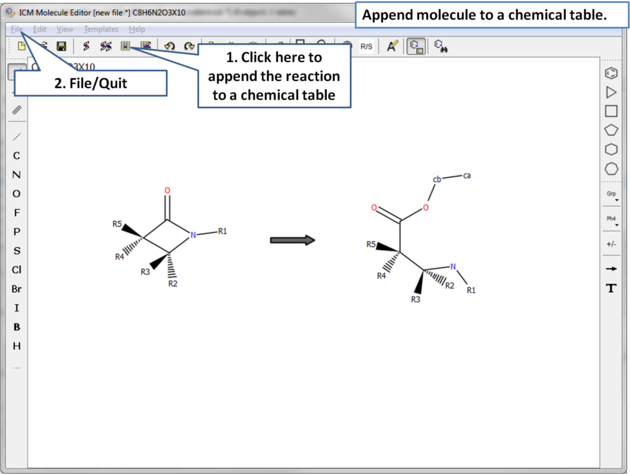 |
| Append the reaction to a chemical spreadsheet (table). If you want to view the atom labels (ca, cb) please select the mol column and choose "Chemical View Options" > "Show Full Atom Names". |
You can check if your reaction matches the chemical(s) you are going to dock by:
- File/Open and read in the chemical sdf file you are going to dock.
- Right click on the reaction you sketched and choose "Apply Reaction".
- Select the chemical table of the chemicals you are going to dock.
- Click on enumerate.
- If the reaction matches the chemicals you are going to dock then you should see a new table called T_react.
Once the reaction has been sketched you can start the covalent docking procedure.
| Prev Template Docking | Home Up | Next AI Generator |