Google Search: Keyword Search:
| Prev | ICM User's Guide 7.1 Making Slides | Next |
| Available in the following product(s): ICM-Browser | ICM-Browser-Pro | ICM-Pro |
A slide enables you you to store a large number of different 3D visualization along with text and window layout.The following information can be stored in a slide:
- Viewpoint (e.g. rotation, translation, zoom, lighting and depth effects)
- A set of atom-specific graphical representations such as surfaces, which can be represented as smooth, transparent or wireframe, ball-and-stick models, CPK (space-filling) models.
- A set of atom, residue or distance labels on any of the atomic items
- A set of arbitrary 3D textual annotations assigned to a point in space
- A set of arbitrary 2D annotations assigned to specific 2D coordinates on a screen
- Parameters of the parametric animation
- Window layout - for when the slides are viewed in the browser.
- Current table(s)
- Sequences and Alignments
- HTML text with hyperlinks
- 2D images
- For each grob (mesh): representation and colors.
A slide can be made by clicking on the Camera button at the bottom of the GUI.
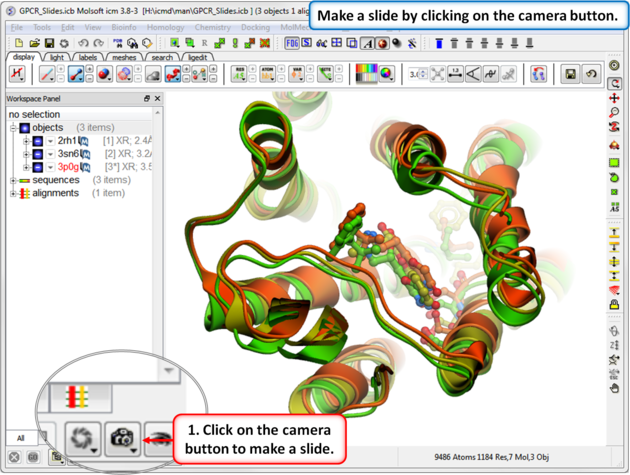
Slides can be viewed in the GUI, can be edited and graphical effects can be added to them. Slides can also be embedded into the web, or PowerPoint using ActiveICM or viewed in ICM-Browser (or ICM-Pro).
This tutorial takes you through the steps to create a series of fully interactive 3D slides. Another tutorial can be found here. To begin making ICM Molecular Slides:
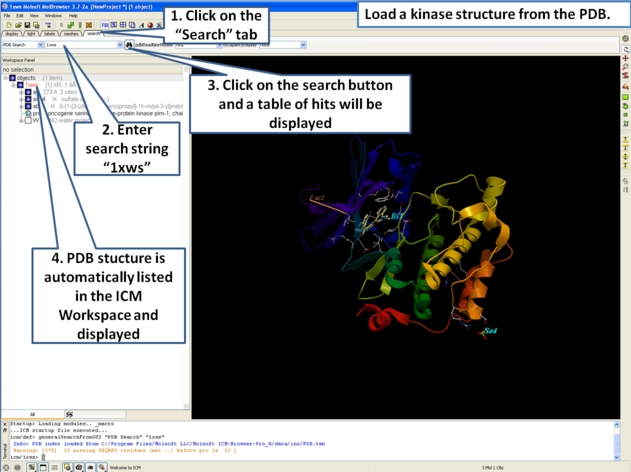
- First load the structure or structures you wish to display in your first slide. Additional structures, labels etc and text can be added at any point during the slide making process. In this example we will load the PDB file 1XWS a PIM1 kinase.
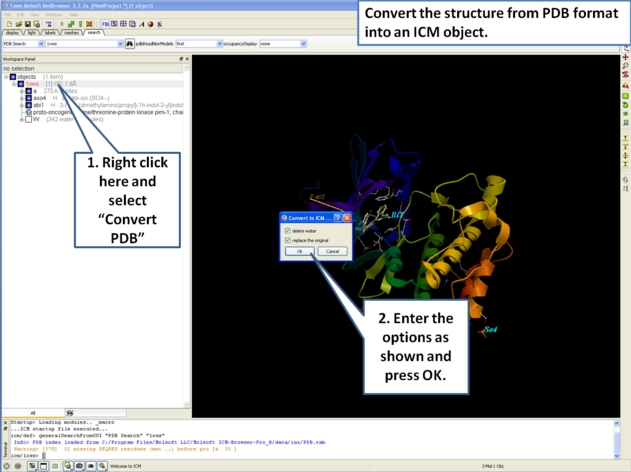
- Next, we will convert the PDB file to an ICM object so we can make slides of the ligand-receptor hydrogen bonds and binding pocket surface.
- Now we are going to prepare the first slide by rotating the protein structure to an orientation which allows the viewer to see the key features of the kinase. For example the bulge in the hinge region (between the N- and C- lobes) which is unique to PIM proteins.
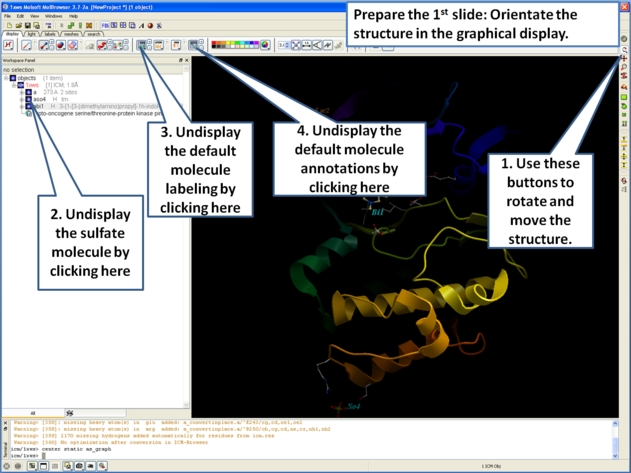
- Next, make the first slide by clicking on the camera button at the bottom of the graphical user interface.
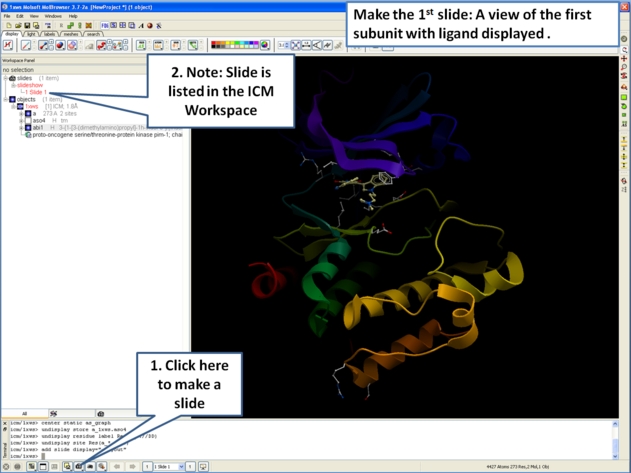
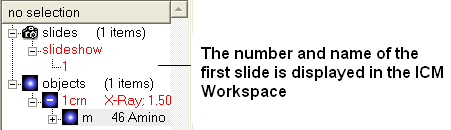
- Once you have clicked on the camera button you will see that the first slide has been generated. The first slide is shown in the ICM Workspace window as
shown below.
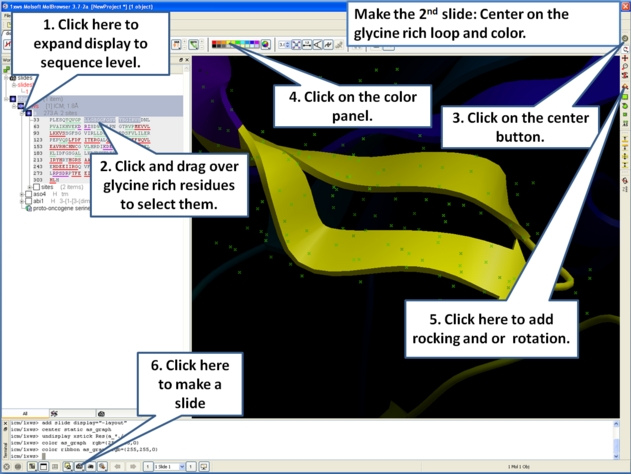
- Slides can consist of Static views or Transitions and Animations. Here we will zoom into the flexible glycine rich region of the kinase which lays across the roof of the ATP-binding pocket. Click on the camera button and make the second slide
- Next, we will make a slide of the surface of the ligand binding pocket colored by binding property.
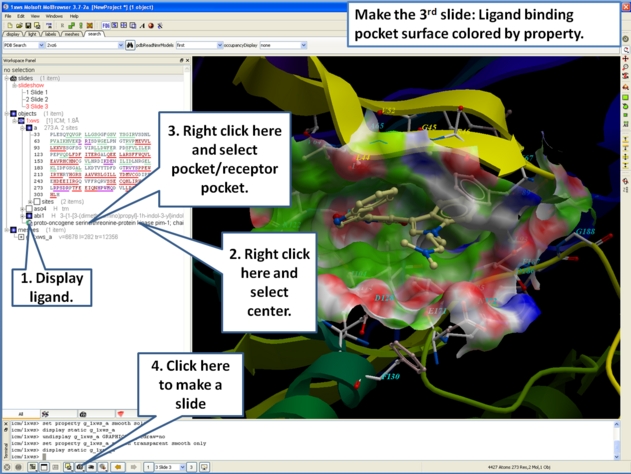
- Now save the document as an icb file. Go to File/Save as...
| Prev Slides & ActiveICM | Home Up | Next Slide Movie |