Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 4.2 How to search and download Protein Structure, Sequences, and Chemicals | Next |
[ Search PDB | Pocketome | ChEMBL | SureChEMBL | BLAST | UniProt | Ligand Code | Drug Bank | Search PubChem | Historeceptomics ]
| Available in the following product(s): ICM-Browser | ICM-Browser-Pro | ICM-Pro | ICM-Chemist |
The Search tab allows you to search and download content from the following databases:
- The Protein Databank www.wwpdb.org - search for protein and chemical
- The Pocketome Database http://pocketome.org/
- BLAST search the NCBI sequence database
- The UniProt Sequence database http://www.uniprot.org/
- Search the PDB by ligand code.
- Search Drug Bank http://www.drugbank.ca/
- Search PubChem https://pubchem.ncbi.nlm.nih.gov/
- Search ChEMBL https://www.ebi.ac.uk/chembl/
- Search SureChEMBL https://www.surechembl.org/search/
- Search the Crystallography Open Database http://www.crystallography.net/
- Search the HistoReceptomics data from GeneCentrix http://www.historeceptomics.com/
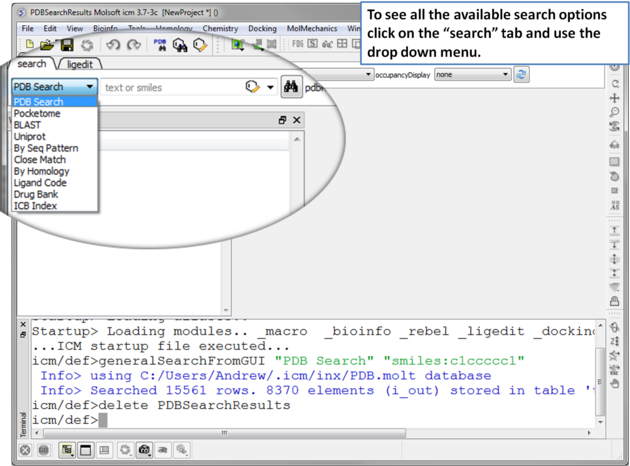 |
| Click on the search tab and then use the drop down menu to identify the database you would like to search and download from. |
4.2.1 Search the PDB |
[ Chemical Substructure | Query Sequence | PDB Field | Load NMR | Occupancy Display | Header | PubMed ]
How to Search the Protein Databank and Download
The PDB search tab provides easy access to the PDB database. You can use keyword searching or type in the PDB code you are interested in. An asterisk (*) wildcard can be used to list all the pdb files currently available in the protein databank. Different fields can be searched by using the drop down arrow as shown below. More advanced PDB search tools and how to use the PDB search result table are described in the section entitled Searching the PDB.Once a search is complete a table of PDB files relating to your search query will be displayed. To view the PDB file in 3D in the graphical display double click on a row in the PDBSearchResults table.
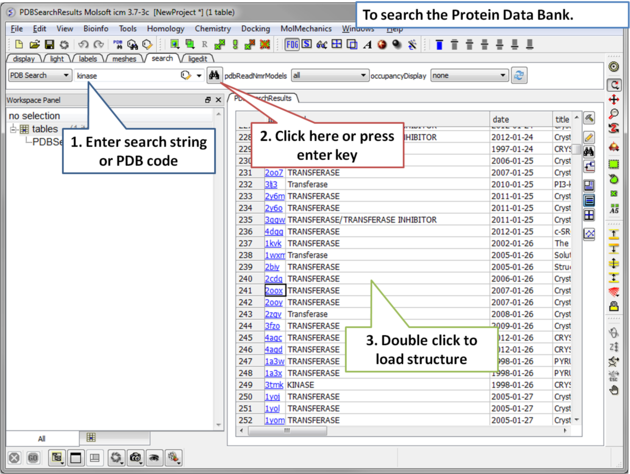 |
To Search the PDB and Download a Structure:
|
| NOTE: If you have a PDB structure already saved you can read it into ICM by going to the File Menu and selecting Open. PDB files that have been viewed previously can be loaded using File/ Recent PDB Codes. |
4.2.1.1 How to Search the Protein Databank by Chemical Substructure |
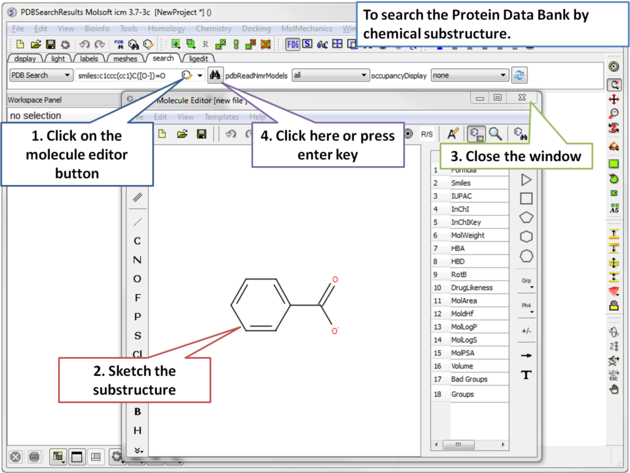 |
| Step 1: Click on the molecule editor button inside the search tab. Sketch the substructure you are interested in. |
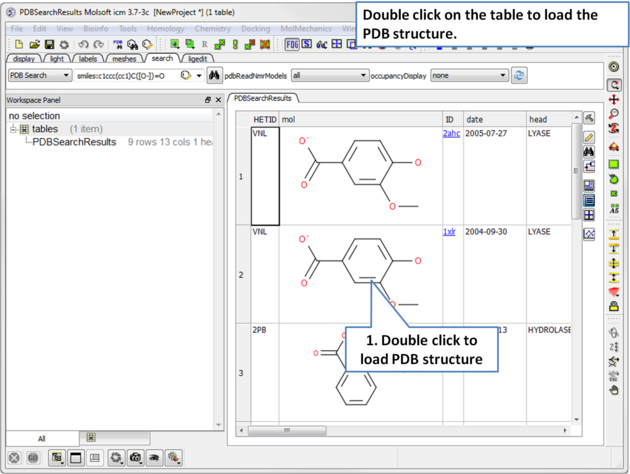 |
| Step 2: A table containing the results will be displayed. Double click on a row to load the PDB file. |
4.2.1.2 How to Query the PDB by Sequence |
There are a number of ways to search the PDB by sequences.
- BLAST search the NCBI datase using the option "Sequences from PDB".
- Search by Sequence Pattern using the option in the Edit menu.
- Search by Identity using the option in the Edit menu.
- Search by Homology using the option in the Edit menu.
4.2.1.3 How to Query PDB by PDB Field |
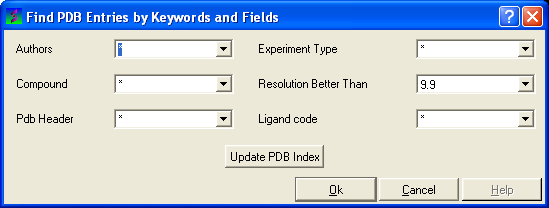
To query the PDB by field (Author, Compound, PDB Header, Experiment Type, Resolution or Ligand Code
- Select Edit/PDB search by field.
- Enter the search string or value
- Click OK and a list of related PDB entries based on your search will be displayed in the PDBSearchResults table of the graphical user interface.
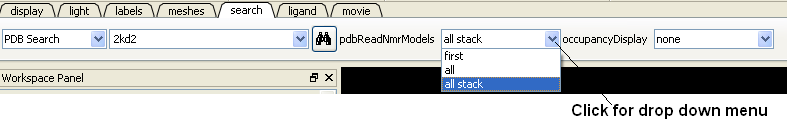
4.2.1.4 Load and Display NMR Structures |
Use the PDB Search tab to load NMR structures from the PDB. You can use the drop down button shown below to determine how you want to display your NMR structure. You can choose to display and download the first NMR model, all models in the PDB file or all models in the PDB in a stack. If you choose the stack option the the stack will be stored in the object as described here.
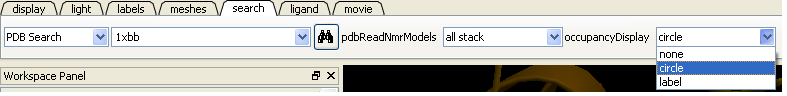
4.2.1.5 Occupancy Display |
You can use the options in the PDB Search tab to control if and how the partial or zero atom occupancies are displayed. You can choose to circle or label the poor occupancy atoms.
>>load-pdb-hyperlinks{pdb search hyperlinks} h4-- Hyperlinks to PDB Website and UniProt {Hyperlinks to Databases}
In the PDB Search Results Table you will see blue hyperlinks that will take you directly to the PDB website or Uniprot website.
4.2.1.6 Display PDB Header |
To display the PDB Header for a PDB file.
- First load a PDB file into ICM (see Search PDB)
- Double click on the word header in the ICM Workspace.
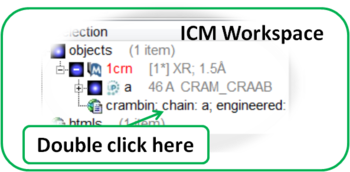
- The PDB Header information will be displayed.
- Click on the blue hyperlinked text to link to external web pages for additional information if needed.
4.2.1.7 Direct link to PubMed |
When you search for a PDB file and load it into ICM you will see an icon (shown below) next to your protein name in the ICM Workspace. Click the icon and you will be taken directly to the PubMed primary reference relating to the structure.

4.2.2 Search Pocketome |
The Pocketome (www.pocketome.org) is an encyclopedia of conformational ensembles of all druggable binding sites that can be identified experimentally from co-crystal structures in the Protein Data Bank.
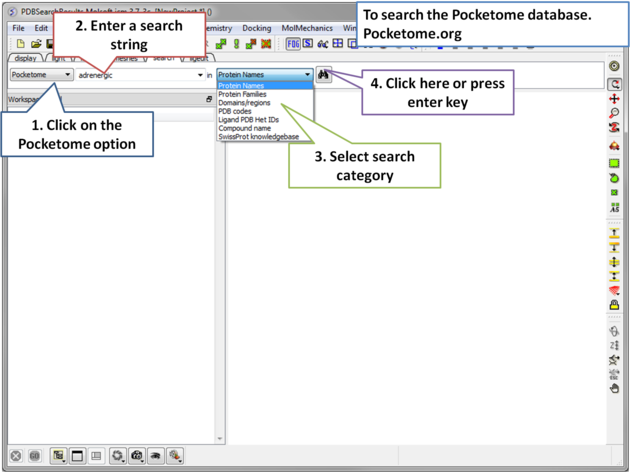 |
| Step 1: Click on the search tab and choose the Pocketome option. Check a field you would like to search e.g. Protein Name, Family, domain... |
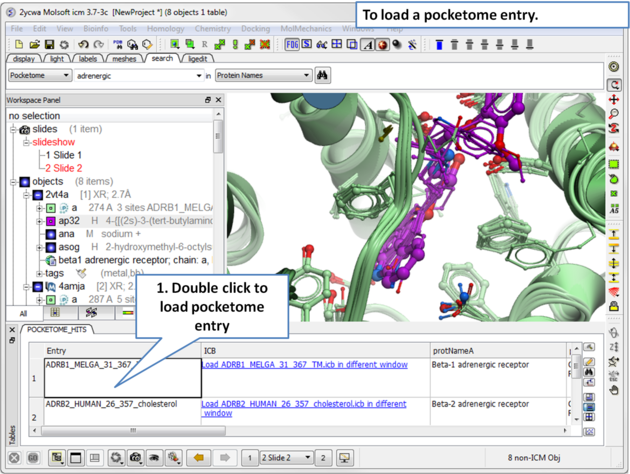 |
| Step 2: A table of pocketome hits will be displayed. Browse the results and double click to load a pocketome entry. |
4.2.3 ChEMBL Search |
ChEMBL (https://www.ebi.ac.uk/chembl/) is a manually curated chemical database of bioactive molecules with drug-like properties. It is maintained by the European Bioinformatics Institute (EBI), of the European Molecular Biology Laboratory (EMBL), based at the Wellcome Trust Genome Campus, Hinxton, UK.
To search and download data from ChEMBL:
- Click on the Search tab.
- Choose ChEMBL from the drop down button.
- Enter a search string or search by chemical sketch by clicking on the editor button in the panel.
- Select a field to search e.g. Protein Name.
- Select whether you wish to return just activity data or all data.
- Click the search button.
A table will be displayed as shown below.
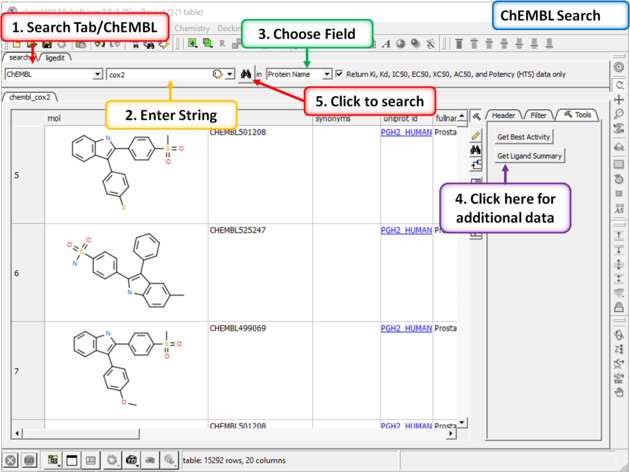
The table contains the original activity data without merging or aggregation.
For instance,
Chemical 1, Protein 1, Activity 1
Chemical 1, Protein 1, Activity 2
Chemical 1, Protein 2, Activity 1
Chemical 2, Protein 1, Activity 1
Click on the button Get Best Activity to display the best activity for each chemical, protein pair.
For instance,
Chemical 1, Protein 1, Best(Activity 1, 2, 3, ...)
Chemical 1, Protein 2, Best(Activity 1, 2, 3, ...)
Chemical 2, Protein 1, Best(Activity 1, 2, 3, ...)
Click on the button Get Ligand Summary to display a list of chemicals with all activities found in ChEMBL.
For instance,
Chemical 1, Protein 1, Activity 1
Protein 1, Activity 2
Protein 2, Activity 1
Protein 3, Activity 1
Chemical 2 Protein 1, Activity 1
Protein 2, Activity 1
Protein 2, Activity 2
4.2.4 Search SureChEMBL |
SureChEMBL (https://www.surechembl.org/search/) provides free access to chemical data extracted from the patent literature. To search SureChEMBL:
- Click on the Search tab.
- Choose SureChEMBL from the drop down button.
- Enter a search string or search by chemical sketch by clicking on the editor button in the panel.
- Select a field to search e.g. Protein Name.
- Select whether you wish to return just activity data or all data.
- Click the search button.
4.2.5 BLAST Search |
To BLAST search the NCBI sequence database:
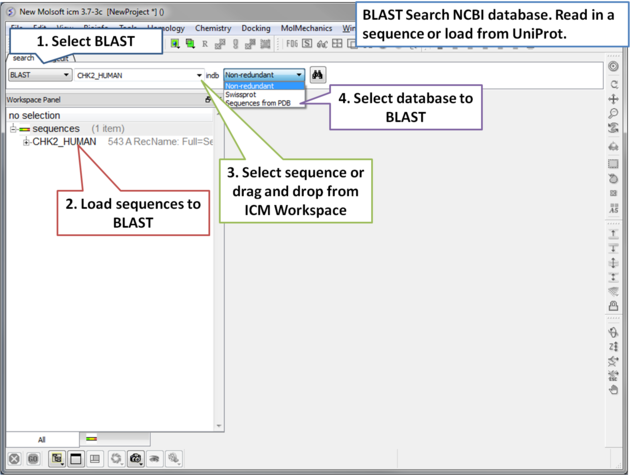 |
| Step 1: Load a sequence into ICM. Select the Search tab and choose the BLAST option. Drag and drop the sequence into the search field, use the drop down menu or type the sequence name. |
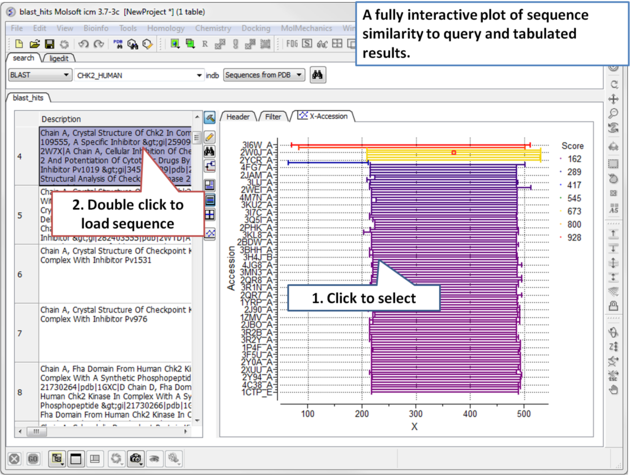 |
| Step 2: A fully interactive table and plot of sequence conservation will be displayed. Double click to load a sequence. |
4.2.6 Search UniProt |
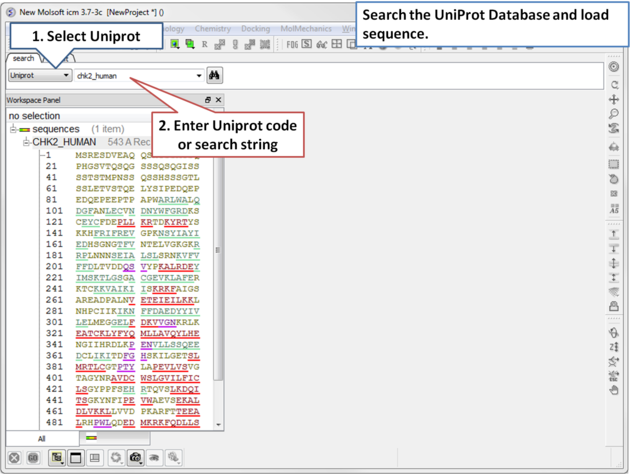 |
| Step 1: Click on the search tab and select Uniprot from the drop down menu. Enter the UniProt code. The sequence will be loaded directly into ICM and you will see it in the ICM workspace. |
4.2.7 Search PDB by Ligand Code |
To find structures containing a particular ligand in the PDB.
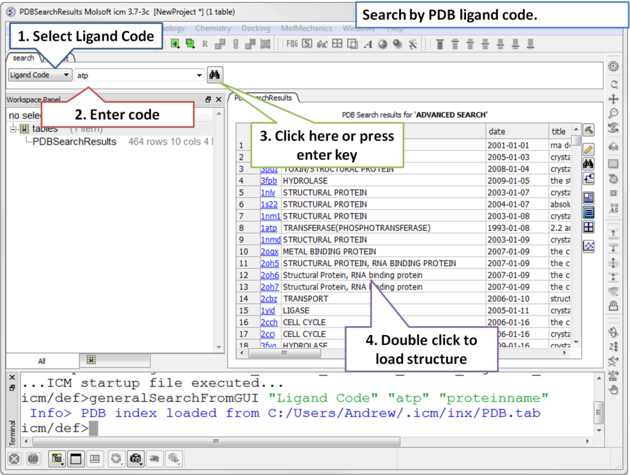 |
| Step 1: Click on the search tab and select Ligand Code from the drop down menu. Enter the Ligand code and a table of hits will be displayed. |
4.2.8 Search Drug Bank |
To search Drug Bank ( http://www.drugbank.ca/)
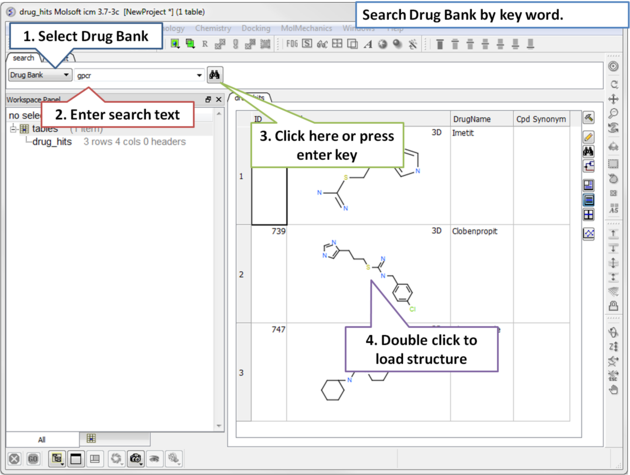 |
| Step 1: Click on the search tab and select Drug Bank from the drop down menu. Enter a search string and a table of results will be displayed. |
The table contains the original activity data without merging or aggregation.
For instance,
Chemical 1, Protein 1, Activity 1
Chemical 1, Protein 1, Activity 2
Chemical 1, Protein 2, Activity 1
Chemical 2, Protein 1, Activity 1
Click on the button Get Ligand Summary to display a list of chemicals with all activities found in Drug Bank. The button is located in the extra panel (click on hammer - top right of table panel).
For instance,
Chemical 1, Protein 1, Activity 1
Protein 1, Activity 2
Protein 2, Activity 1
Protein 3, Activity 1
Chemical 2 Protein 1, Activity 1
Protein 2, Activity 1
Protein 2, Activity 2
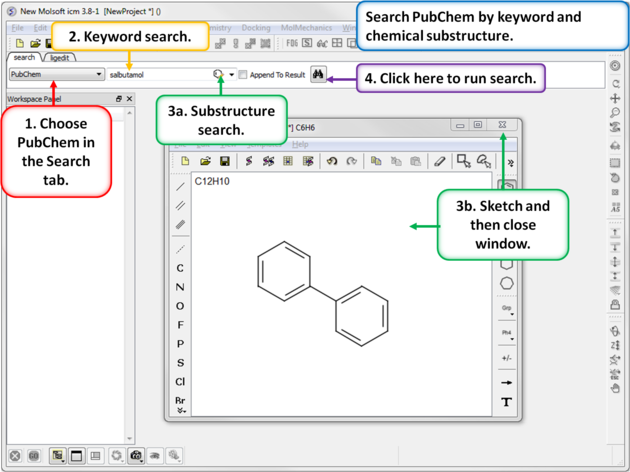
4.2.9 Search PubChem |
To search PubChem (https://pubchem.ncbi.nlm.nih.gov/):
 |
|
4.2.10 Historeceptomics |
The historeceptomics (HR) platform is developed by GeneCentrix and reports tissue specific profiling for a drug molecule. It is a unique bioinformatics platform designed to identify mechanism of activity and predict unforeseen effects a chemical might have in the target tissue and off-target tissues.
HR integrates data from a variety of sources including:
- BioGPS
- GTex
- ChEMBL
- DrugBank
- BindingDB
- Genecentrix database
- Select the Search tab.
- Select the historeceptormics option from the drop down search menu.
- Enter a drug name or sketch a chemical using the molecular editor.
- Choose the BioGPS or GTex database. Selecting BioGPS will use the GeneAtlas U133A dataset from BioGPS to analyze the tissue-wide expression profiles of targets of interest. Selecting GTEx will use the median gene-level TPM by tissue dataset from GTEx to analyze the tissue-wide expression profiles of targets of interest
- Click the search button.

After searching a results panel as shown below will be displayed.
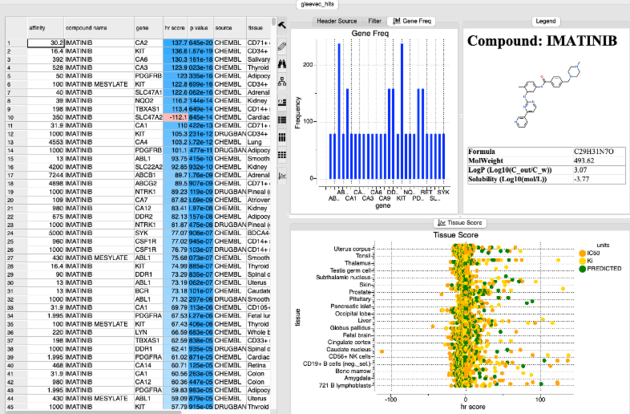
The table is ranked by hr Score and reports the binding affinity gene name and tissue. The p-value represents the chance that the overabundance of the tissue/target in question could occur by random chance based on the variability of the expression of that target across 70 other tissues. Additional plots of gene frequency and hr score for each tissue are reported.
| Prev Pictures | Home Up | Next Create New Objects |