Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 10.9 Find Bioisostere | Next |
[ 2D Bioisostere | 3D Bioisostere ]
In drug design bioisosteres can be used to reduce toxicity, change bioavailability, alter metabolism and change activity of lead compound. Bioisosteres are chemical substituents with similar chemical or physical properties which produce broadly similar biological properties to another chemical compound.
10.9.1 2D Bioisostere |
To find 2D Bioisosteres:
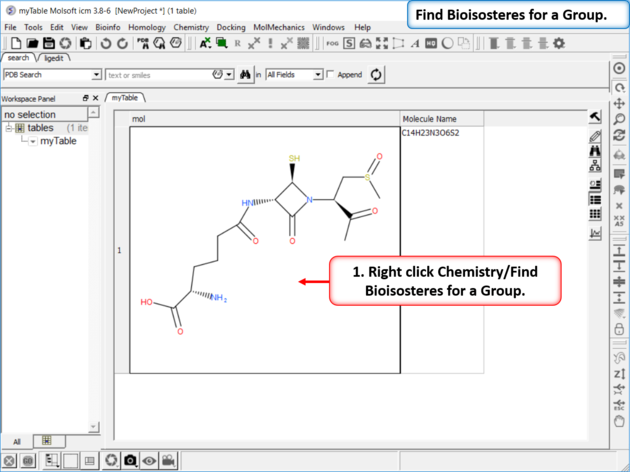 |
| Step 1: Read in a chemical into a chemical table. Right click Chemistry/Find Bioisosteres for a Group. |
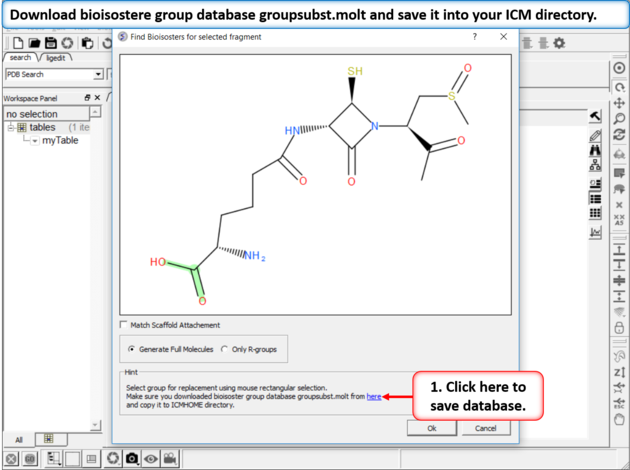 |
| Step 2: Download Database If you do not see this link it means you already have the database in your ICM directory. If yo do not have it - download bioisostere group database groupsubst.molt and save it into your ICM directory. There is a link at the bottom of the dialog window which will direct you to the correct place to download. |
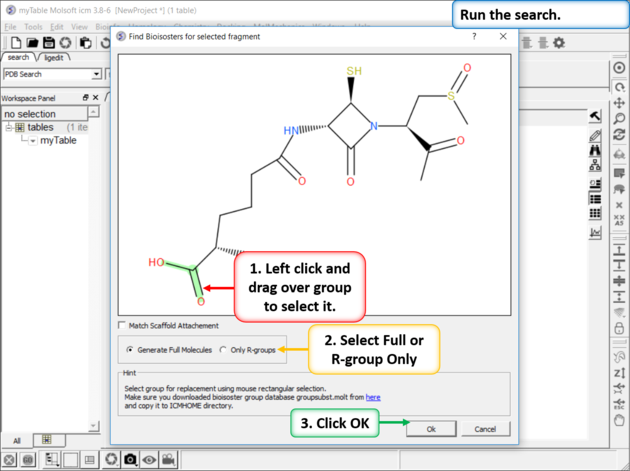 |
Step 3: Setup the Search
|
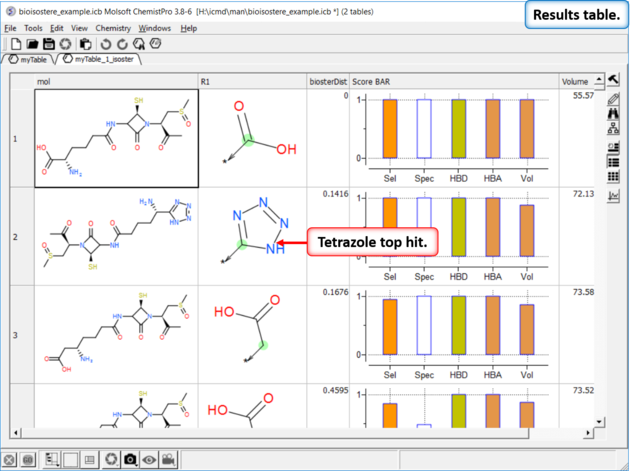 |
Step 4: Results Table
In this example we find tetrazole which is a classic isostere of carboxylic acid. The data in the columns represent:
|
If you double click on a row in the table it will show specific examples of a particular substitution from ChEMBL.
10.9.2 3D Bioisostere |
The receptor/ligand complex structures from the PDB contains relevant (bioactive) 3D conformation and the environment of the isosteric moieties. This can be used to find 3D bioisosteres. You can watch a Bioisostere Webinar which explains how the 3D database was constructed.
To find 3D Bioisosteres:
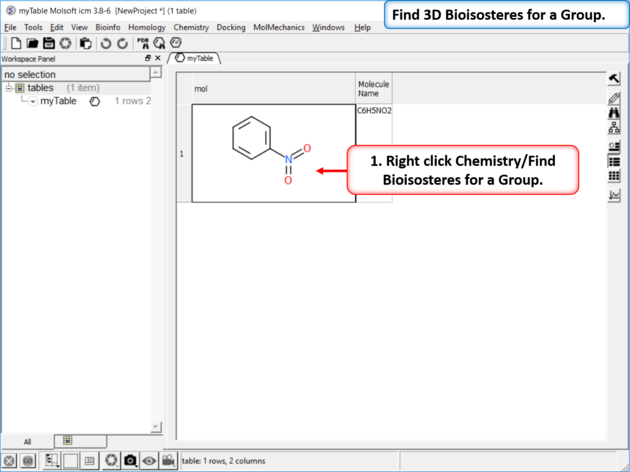 |
| Step 1: Read in a chemical into a chemical table. Right click Chemistry/Find Bioisosteres for a Group.
|
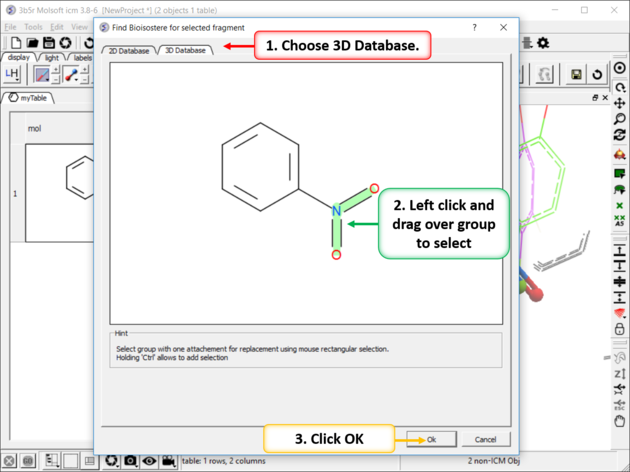 |
| Step 2: Left click and drag over the group you wish to find a 3D bioisostere for and then click OK. |
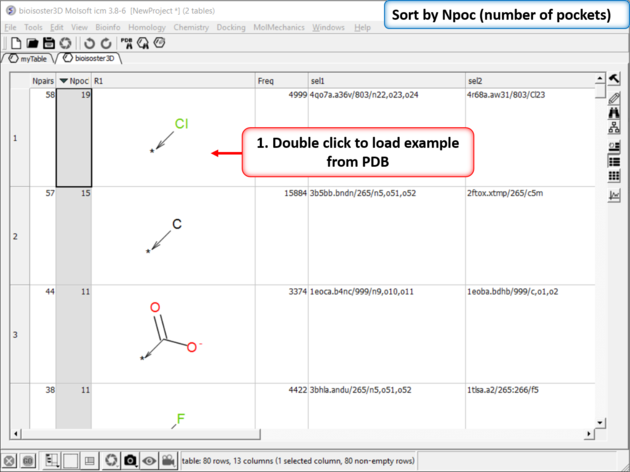 |
| Step 3: The results table will be displayed the nPocs (number of pockets) column is a good indicator of a good bioisostere. As this is the number of times this replacement is seen in the PDB. |
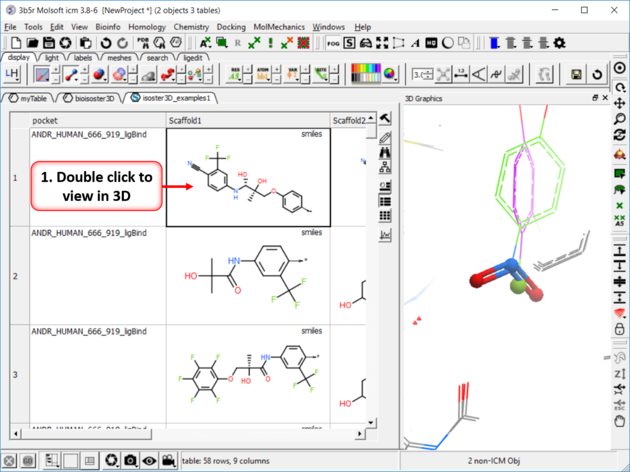 |
| Step 4: Click on any of the hits to view specific pair examples in 3D. |
| Prev Fragments | Home Up | Next Molcart |