Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 12.12 Peptide Docking | Next |
|Video Example|
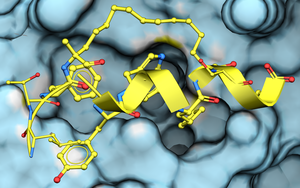
To dock a peptide to a protein structure:
1. Setup the Docking Project and Receptor
- Setup the protein receptor the same way you would for small molecule docking.
- Prepare a table with your peptide sequence(s) to dock from Docking/Dock Peptide table.
2. Preparing a Table with Peptide Sequences
To create a table for peptide sequences, follow these steps:
Create a New Table
- Go to File > New and select the Table tab.
- Choose one string column and set the number of rows based on how many peptides you wish to dock.
- A new table will appear in the GUI.
- Rename the column containing the sequence to 'sequence' .
- Enter your peptide sequences in the format shown below:
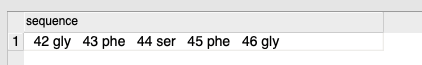
Sequence Format Guide
Before defining sequences, check icmff.res to see which residue types ICM supports. All defined residues are stored in the icmff.res file in the ICM distribution directory.
It is a text file so you can view it in a text editor or grep it Example to search for ornithine grep ornithine icmff.res and you will see the code is 'orn'
1. Standard and Advanced Numbering
- 3-Letter Codes:
gly ala ser pro tyr his - Advanced Notation: (Includes numbering, N/C-terminals, and D-amino acids)
0 nter 1 gly 2 ala 2A Dglu 4 asp cooh
2. Specialized Bonds and Cyclization
- Disulfide Syntax: Use numerical markers in parentheses. Multiple disulfides can be encoded as (1), (2), etc.
acet ala cys(1) ala ala cys(1) ala conh - N-term to C-term Cycles: Use special "virtual" residues:
nvtr ala ala ala ala cvtr
3. Crosslinks and Modifiers
Use curly braces {} for crosslinks or unusual modifications.
- Lys to Glu Link: The 'Xa' stands for crosslink 'a' replacing an atom (e.g., Nz of lysine).
acet ala lys{nz_Xa} ala ala ala ala ala gln{he22_Xa} conh - Unusual Residues:
aba{hg1_C} ser{hg_C(=O)C)}
Note: Modifiers cannot be applied to the backbone (C, Calpha, and N atoms). Hydrogen and side-chain atoms are acceptable.
4. Terminal Designations
| Terminal Type | ICM Residue Code |
|---|---|
| C-terminal amide | conh |
| N-terminal charged amine | nh3+ |
| N-terminal neutral amine | nter |
Example Input File Sequence:
nh3+ nle ala ala ala ala conh
Importing Sequences (Optional)
- You can import a table from an Excel or CSV file. Make sure that the column containing the sequence is labeled 'sequence'.
- If you have a peptide loaded from a PDB file, extract its sequence:
- Right-click on the peptide.
- Select>Extract Sequences to Table.
3. To dock the ligand:
Docking a Peptide Table
Start Docking
- Setup the protein receptor the same way you would for small molecule docking.
- Go to Docking > Dock Peptide Table.
- Click OK to use the default docking settings.
Adjusting Docking Parameters (Optional)
- If you uncheck "dock immediately," you can modify settings before docking.
- Use the table side panel to adjust:
- Thoroughness
- Number of conformations
Restraining or Biasing the Peptide Structure
You can restrain or bias the ligand towards a specific secondary structure. To do this, define each residue with one of the following secondary structure types:
- H - Alpha helix
- G - 3/10 helix
- I - Pi helix
- E - Beta strand
- B - Beta-bridge
- _ - Undefined
- C - Coil
| Prev Autofit | Home Up | Next Protein-Protein Docking |