| Prev | ICM User's Guide 10.2 Quick Superimpose | Next |
In order to calculate the root mean square deviation (RMSD) between two structures it is necesary to superimpose them. By using the superimpose button in the display tab, ICM will calculate the Ca-atom, backbone atom and heavy atom differences between the two structures. More advanced superimpose options can be found in the Tools/Superimpose menu.
To superimpose two structures which have the same number of residues and atoms:
- First load the two structures into ICM.
- Select which parts or all of the two structure you wish to superimpose (see selection toolbar).
- Select the display tab (previously called Advanced tab) at the top of the GUI.
- Select the superimpose button.

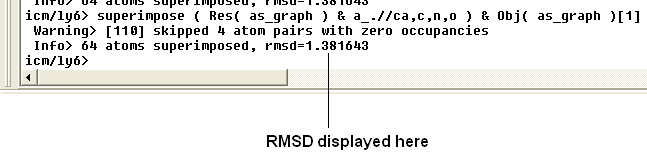
The rmsd will be displayed in the terminal window as shown below:
| Prev Display and Select Proteins for Superposition | Home Up | Next Proteins by 3D |