| Prev | ICM User's Guide 10.4 Superimpose Multiple Proteins | Next |
To superimpoe multiple proteins:
- First display and select the proteins you wish to superimpose by 3D.
- Tools/Superimpose/Multiple Proteins
- A window as shown below will be displayed.
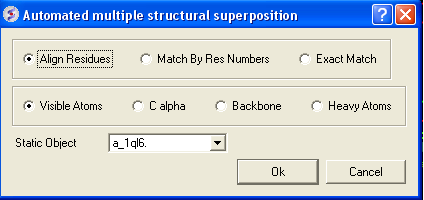
- Select by which method you would like to superimpose
Align Residues - Residue correspondence is established by sequence alignment using the ICM ZEGA alignment Abagyan, Batalov, 1997. Atom alignment: by atom name.
Match by Res Numbers - Residue alignment by residue number.Atom alignment: by atom name for pairs of identical residues or pairs of close residues (F with Y; B with D,N; D with N; E with Q or Z, Q with Z), for other residue pairs only the backbone atoms ca,c,n,o,hn,ha are aligned.
Exact Match - Residue alignment is by the Needleman and Wunsch method. Inside residue atoms are aligned sequentially and regardless of the name.
- Select which atoms you would like to superimpose. Visible Atoms, C alpha, Backbone, or Heavy Atoms.
| Prev Proteins by 3D | Home Up | Next Arrange as Grid |