| Prev | 3.2 Making Selections | Next |
[ Graphical | Selection | Basic | Altering | Filter | Workspace | Workspace | Whole | Amino | Neighbors | Neigh/Graphical | Neigh/Workspace | Alignment and Table | Links ]
| Note: Click Next (top right hand corner) to navigate through this chapter. Headings are listed on the left hand side (web version) or by clicking the Contents button on the left-hand-side of the help window in the graphical user interface. |
There will be many occasions when you will have to make selections. For example, if you only wish part of your structure to be displayed or if you want to select residues around a binding pocket. If you have a molecule displayed in the graphics window, then selections will be displayed as green crosses. The selection you have made is also displayed at the top of the ICM Workspace. It is always a good idea to keep an eye on what is selected and what isnt.
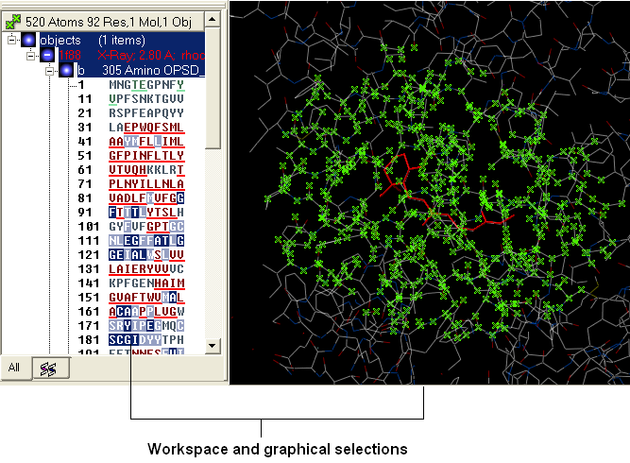
There are four basic levels of selection
- Object (eg a PDB structure or ICM object)
- Molecule
- Residue
- Atom
You can make selections in:
- The Graphics Display
- The ICM Workspace (Selections are highlighted in blue)
- Tables
- Alignments
3.2.1 Graphical Selections |
In this section you will learn how to select parts and certain regions of molecules from the 3D graphical display. Graphical and molecule selections are required in many operations within ICM. For example, if you wanted to display graphically part of a molecule or if you wanted to perform a minimization of residues within a sphere of an imporant atom.
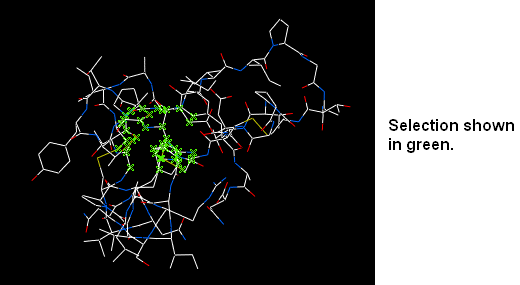
3.2.2 Selection Tools |
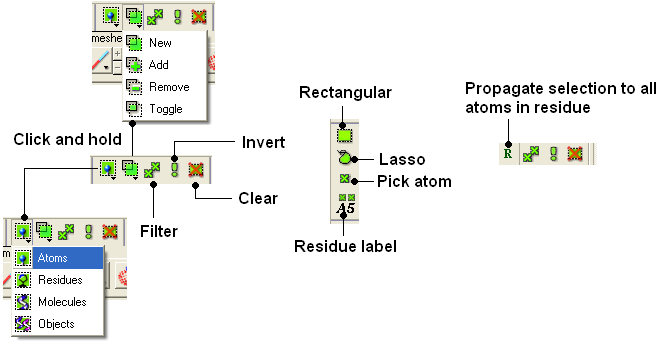
The following buttons can be used to make a selection once a structure is displayed.
| NOTE: All selection tool buttons are colored green. Graphical selections are represented as green crosses. |
3.2.3 Basic Selections |
To make a basic selection (ie nothing too complicated!) the following buttons can be used.

To unselect everything you have previously selected:
- Simply click on the Unselect icon on the selection toolbar.
OR
- Right click and drag away from the displayed structure.
To select parts of your structure:
- Click on the Rectangular selection icon and click and drag around the part of the structure you wish to select.
OR
- Click on the Lasso selection icon and click and drag your mouse around the area of the structure you wish to select, forming a lasso around it.
To pick individual atoms:
- Click on the 'pick atom' button
You can also change the level of a selection using the button shown below. Click and hold the button to choose the level of selection. For example, if you have selected atoms you can convert the selection to all atoms at the residue level by choosing the Residues option.
- Click on the Select objects , Select molecules, Select residues, or Select atoms icon, depending on which part of the structure you wish to be highlighted.
| NOTE: The selection you have made is always recorded at the top of the ICM workplace. If you are familiar with using the ICM terminal (See language manual) the atoms, residues, molecules or objects selected interactively in the graphics window are automatically stored in the as_graph variable. |
3.2.4 Altering a Selection |
Once you have made a selection you may wish to add or remove parts of the selection. The buttons shown below allow you to accomplish this.
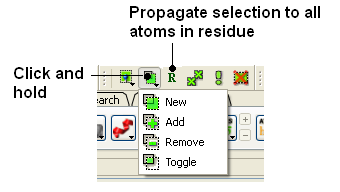
To add or remove from your current selection:
- Click on the Selection mode: add or Selection mode: remove icon on the toolbar.
- Click and drag around the part of your structure you wish to add or remove.
You may also wish to invert your selection in a specific part of the structure.
The parts that are currently selected will become unselected, and the unselected parts will become selected.
In order to invert a selection:
- Click on the Invert icon on the toolbar.
If you wish to select and unselect certain regions of a selection the toggle selection button is very useful.
- Click on the Toggle selection button.
- Right click around the selections you wish to select or unselect.
| NOTE: The selection you have made is recorded at the top of the ICM workplace. Any selection is stored in the variable as_graph. |
3.2.5 Filter Selection |
You may want to be very specific about a selection you want to make. For example you may only wish to select protein backbone atoms.
The button shown below enables you to filter your selection:

Or
Right click on a selection and a menu as shown below will be displayed.
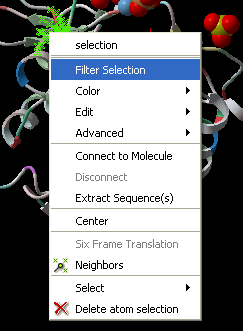
- Select the Filter Selection option.
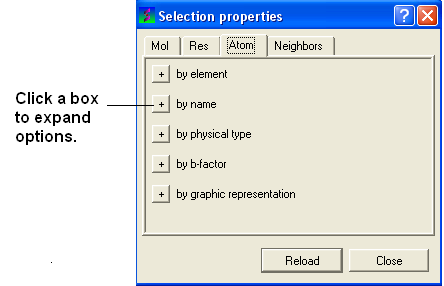
If you wish to filter and select by residue or atom type:

- Click on the Filter graphical selection icon on the toolbar and a data entry box as shown below will be displayed.
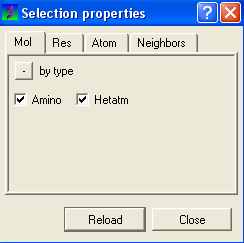
To select just the protein or just the hetatoms as well:
- Click on the Mol tab.
- Check the appropriate boxes depending on your desired selection.
To filter by residue type or secondary structure:
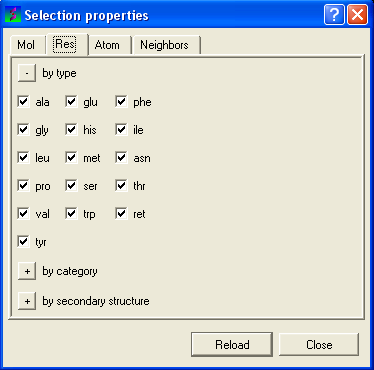
- Click on the Res tab.
- Check the appropriate boxes.
| NOTE: You may need to click on the button marked with a '+' symbol to expand the options. |
To filter by atom type.
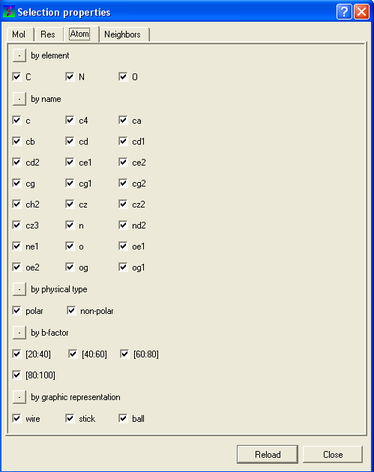
- Click on the Atom tab.
- Check the appropriate boxes.
| NOTE: You may need to click on the button marked with a '+' symbol to expand the options. |
To select neighbors to a particular selection.
- See the select neighbours section for detailed instructions.
| NOTE: The selection you have made is always recorded at the top of the ICM workplace. If you are familiar with using the ICM terminal (See language manual) the atoms, residues, molecules or objects selected interactively in the graphics window are automatically s |
3.2.6 Workspace Selections |
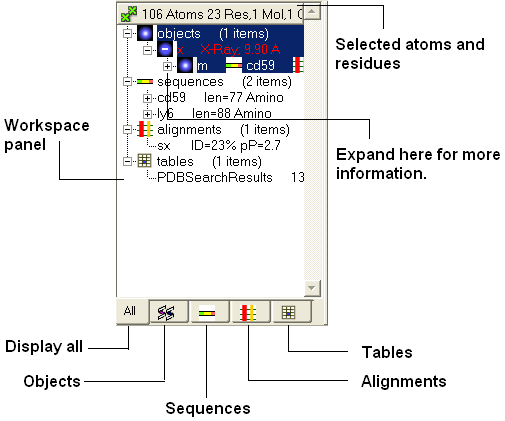
In the default GUI layout the workspace panel is located to the left of the 3D graphics display. It is a great tool for keeping track of all your sequences, pdb structures, objects, tables and alignments. As you will see in this section it also provides a way of making selections.
3.2.7 Workspace Navigation |
Once you have mastered how to navigate the ICM workspace making a selection will become easier. Each object is divided into 3 levels:
- Object Level - Shown in red if it is the current object. Holds details about the structure - name, X-ray, NMR, resolution etc. Importantly it will state whether the structure is an ICM object or a structure straight from the PDB. To learn how to convert a PDB into an ICM object go to the section on converting a PDB.
- Molecular Level - Shows the individual subunits, ligands and hetatoms of a molecule.
- Residue Level - Shows the sequence.
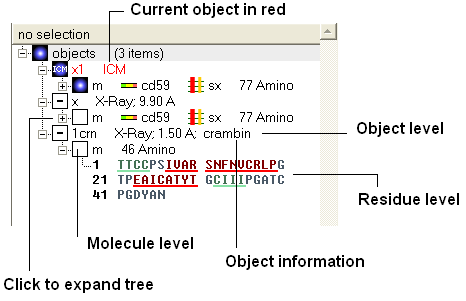
| NOTE: You can expand each level of the ICM workspace by clicking the "+" button as shown above. |
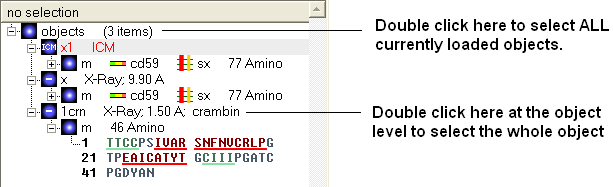
3.2.8 Selecting the Whole Object |
To select the whole object:
- Double click on the object level.
3.2.9 Selecting Amino Acids |
There are three options to select individual amino acid residues:
OPTION 1:
- Click and drag over the residues you wish to select in the ICM workspace. Selected residues will be highlighted in dark blue in the workspace and with green crosses in the graphical display.

OPTION 2:
- Click on the rectangular selection icon or lasso selection icon on the toolbar.
- Click and drag around the residues you wish to select. Selected residues will be displayed by green crosses on the graphical display and blue in the ICM workspace.
- Click on the Pick Atom button.
OPTION 3:
- Right click on the selected residue in the graphical display and a menu as shown here will be displayed.
- Click on Select and a further menu will be displayed.
- Click on Residue, Molecule or Object.
| NOTE: Ctrl + A will select everything in the ICM workspace, and Ctrl + Shift + A will unselect your objects. |
| NOTE: The selection you have made is always recorded at the top of the ICM workplace. If you are familiar with using the ICM terminal (See language manual) the atoms, residues, molecules or objects selected interactively in the graphics window are automatically s |
3.2.10 Selecting Neighbors |
In some instances you may only want to display or select only a subset of a structure. For example you may only wish to display the residues surrounding a ligand (as shown below (ligand red; graphical selection green crosses). The "Selecting Neighbors" option selects the residues within a shpere of a defined radius.
There are two ways of selecting neighbours to a particular atom or residue in ICM. Either by right clicking on the atom or residue in the graphical display or by right clicking in the ICM workspace.
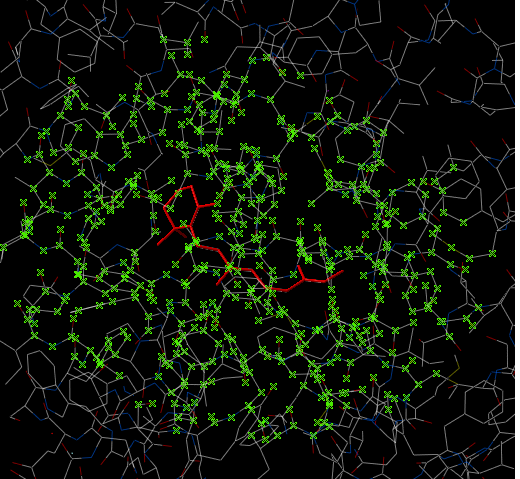
3.2.11 Selecting Neighbors: Graphical |
To select neighboring atoms or residues around a sphere of a certain radius:
- First select the residue(s) or atom(s) around which you wish to select neighbors. (See the Selection Toolbar Section)
- Right click on the selection and a menu as shown below will be displayed or choose Tools/Geometry/Neighbors.
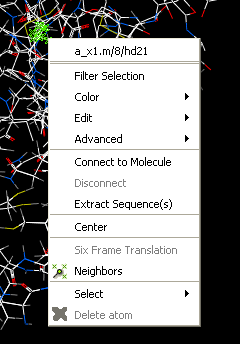
- Select the Neigbors option and a data entry box as shown below will be displayed.
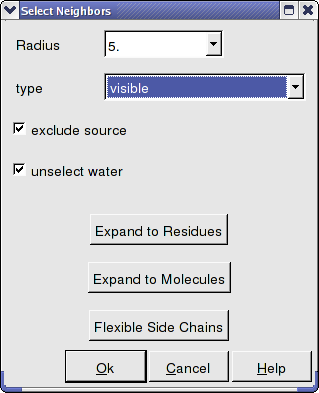
- Enter the selection radius around which you wish to select.
- Select from which object you wish to make the selection from the drop down list in the "type" data entry box.
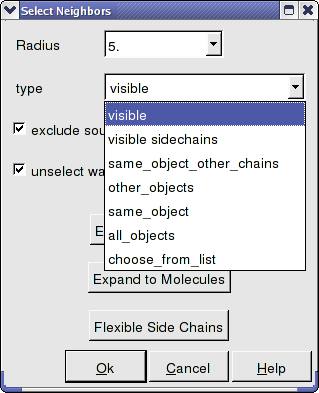
- Select whether you wish your original selection to be selected by checking the "exclude source" box.
- Select whether you wish water molecules to be selected by checking the "unselect water" box.
- There are also options to change the current selection to expand to residues, molecules, or flexible side-chains.
- Click OK and your selection will be displayed as green crosses.
| NOTE: The selection you have made is always recorded at the top of the ICM workplace. If you are familiar with using the ICM terminal (See language manual) the atoms, residues, molecules or objects selected interactively in the graphics window are automatically saved in the variable as_graph. Graphical selections are shown in green (crosses) or highlighted in blue in the ICM Workspace. |
3.2.12 Selecting Neighbors: Workspace |
To select neighboring atoms or residues around a sphere of a certain radius from a residue in the ICM workspace:
- First select the residue in the ICM workspace around which you wish to select neighbors. (See the Residue Selection)
- Right click on the selection and a menu as shown below will be displayed.
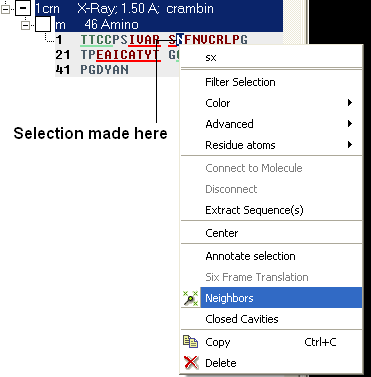
- Select the Neigbors option and a data entry box as shown below will be displayed.
- Follow the instructions in the previous section.
| NOTE: The selection you have made is always recorded at the top of the ICM workplace. If you are familiar with using the ICM terminal (See language manual) the atoms, residues, molecules or objects selected interactively in the graphics window are automatically s |
3.2.13 Alignment and Table Selections |
Descriptions on how to make selections in Alignments and Tables are in the sections entitled Making Selections in Alignments and Making Table Selections.
3.2.14 Making Links |
It is sometimes necesary to make links between sequences objects and alignments. A link enables you to make selections in one environment such as an alignment and then these selections are transfered to the object such as the PDB structure displayed.
If a link is made then a symbol will be displayed next to the object in the ICM workspace. In the example shown below subunit_a of the X-ray structure 1ql6 is linked to the sequence 1ql6_a and the alignment called 'alig'.
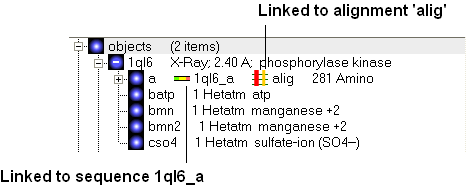
If an object is linked to an alignment a symbol as shown below will be displayed.
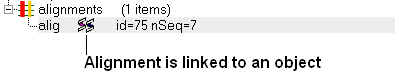
To link a sequence from an object - extract the sequence from the object.
- Right click on the object in the ICM workspace.
- Select extract sequence.
To link a sequence and object to an alignment.
Use the extracted sequence as described above to build your alignment.
In addition a link can be made between a structure and alignment by:
- Bioinfo/Link to Structure.
- Enter alignment name.
- OK
| Prev Pictures | Home Up | Next Menu Option Guide |