| Prev | ICM User's Guide 13.5 Identfy Ligand Pockets | Next |
If a binding pocket is not known in advance, use icmPocketFinder or icmCavityFinder (for closed pockets). The protein needs to be converted to an ICM object in order to use icmPocketFinder.
icmPocketFinder can be accessed by
- Click on the menu Tools/3D Predict/icmPocketFinder
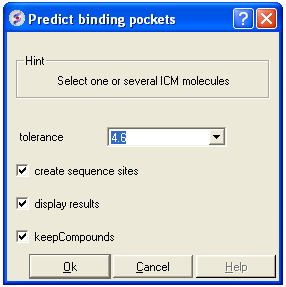
- Enter a tolerance level (4.6 is the default value and we recommended you to use this). The lower the tolerance value the more pockets predicted and the higher the tolerance the less pockets predicted.
- Check the box create sequence sites if you wish the site to be labeled.
- Check the box display results to see the predicted pockets as grobs in the display panel.
- Check the box keep compounds if you wish the compounds (ligands) in the receptor to be included in the prediction. If you dont check this box the pockets will be calculated based on the receptor without ligands.
- Click OK to run icmPocketFinder
| NOTE: A button for icmPocketFinder can be found on the Setup Receptor option in the docking menu. It performs the same function as Tools/3D Predict/icmPocketFinder |
The results from icmPocketFinder will be displayed in a table.
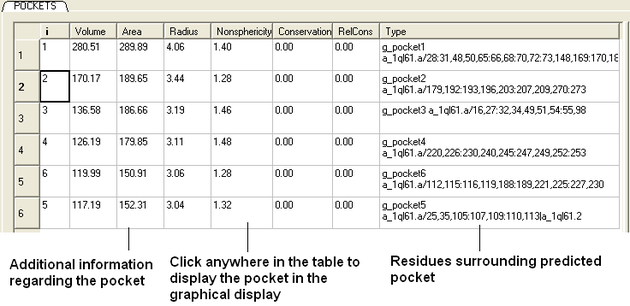
To view the pocket in the graphical display:
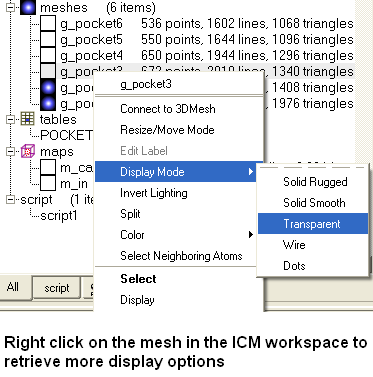
- Click on the pocket in the table or select the pocket from the meshes section of the ICM workspace. Right click on the pocket mesh in the ICM Workspace to retrieve more display options.
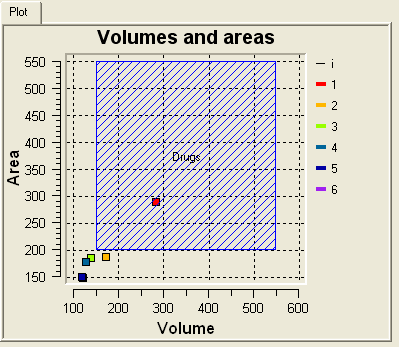
The results from icmPocketFinder are also plotted graphically (Area vs Volume). A blue square highlights potential drug binding pockets based on typical area and volume values - this is only a guide on what constitutes a pocket likely to be involved in ligand binding. Selections can also be made from the plot by clicking and dragging around a point in the graph.
To identify ligand binding pockets which are completely enclosed in the receptor:
- Click on the menu Tools/Analysis/Closed Cavities and a window as shown below will be displayed.
A similar output to that generated by ICMPocketFinder will be displayed. This output includes a plot and a table. By clicking on the table or plot graphical selections can be made.
| Prev Protein-Protein Interface Prediction | Home Up | Next Assign Helices and Strands |