| Prev | ICM User's Guide 8.2 Bioinfo Menu | Next |
[ Content | Secondary Structure | Six Frame Translation | Set Sequence Type | Align Two Sequence | Sequence to Structure Alignment | Align DNA vs Protein | Multiple Sequence Alignment | Link to Structure | Extract Sub-Alignment As Is | Cut Vertical Alignment Block | Reorder Sequences | Extract Unique Sequences | Load Example Alignment ]
| Note: Click Next (top right hand corner) to navigate through this chapter. Headings are listed on the left hand side (web version) or by clicking the Contents button on the left-hand-side of the help window in the graphical user interface. |
8.2.1 Residue Content |
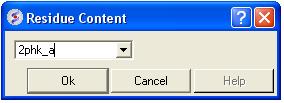
To determine the residue content of a sequence.
- Bioinfo/Residue Content and a data entry box as shown below will be displayed.
- Enter the sequence name. (Go to the Load Sequence section for more information on how to load a sequence into ICM using the Graphical User Interface)
- A table and graph of residue frequencies will be displayed.
8.2.2 Predict Secondary Structure |
To predict the secondary structure of a sequence:
- Bioinfo/Predict Secondary Structure
- Enter the sequence name. (Go to the Load Sequence section for more information on how to load a sequence into ICM using the Graphical User
- An option is provided to ignore currently assigned secondary structure.
To view the secondary structure prediction click on and expand the sequence in the ICM workspace. Regions underlined in red are helices and green represents beta sheet.

8.2.3 Six Frame Translation |
This options returns the translated DNA or RNA sequence ('-' for a Stop codon, 'X' for an ambiguous codon) using the standard genetic code.
- Read into ICM a DNA sequence from a file (eg File/Open FASTA) or use the File/New option and cut and paste a DNA sequence.
- Bioinfo/Six Frame Translation
- Translate all frames or use start codon.
8.2.4 Set Sequence Type |
This option allows you to define whether a sequence that is read into ICM is a protein or nucleotide sequence.
- Read into ICM a sequence (eg File/New and cut and paste sequence or File/Open FASTA)
- Bioinfo/Set Sequence Type
- Select the sequence name using the drop down button
- Select sequence type protein or DNA.
8.2.5 Align Two Sequences |
To align two sequences:
- Read into ICM two or more sequences.
- Bioinfo/Align Two Sequences
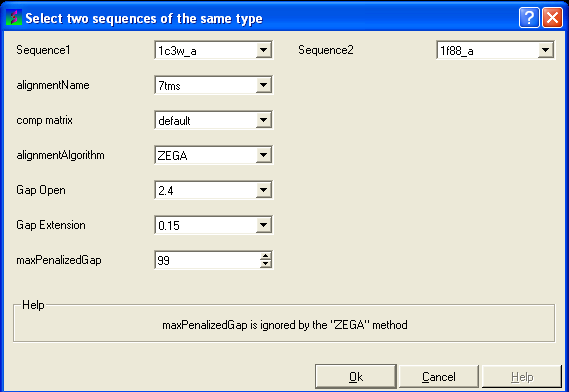
- Enter the name of your first sequence in the 'Sequence 1' data entry box.
- Enter the name of your second sequence in the 'Sequence 2' data entry box.
| NOTE: Any sequences already loaded into ICM can be seen by clicking on the down arrow next to the 'Sequence 1 and 2' data entry boxes. This can save typing and trying to remember what you called your sequence. |
- Enter a unique alignment name in the 'alignmentName' data entry box.
- Select a comparison matrix from the list shown below by clicking on the arrow next to the 'comp matrix' data entry box.
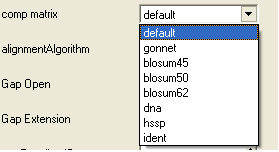
- Select the alignment algorithm you wish to use from the list shown below by clicking on the arrow next to the 'alignmentAlgorithm' data aentry box.
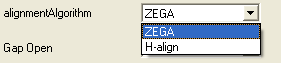
ZEGA - a Zero End-gap Global Alignment, that is a pairwise alignment method based on the Needleman and Wunsch algorithm modified to use zero gap end penalties. This type of alignment was first described by Michael Waterman, who called it the "fit" alignment. The paper of Abagyan and Batalov, 1997 describes the statistics of the structural significance of the alignment score and optimization of the alignment parameters for the best recognition of structurally related proteins.
H-Align - alignment method used in the Align and Score functions and find database command (as described in Batalov and Abagyan, 1999)
- Enter the values you wish to use for Gap Open, Gap Extension and the maximum penalized gap penalty.
Gap Open The absolute gap penalty is calculated as a product of gapOpen and the average diagonal element of the residue comparison table You may vary gapOpen between 1.8 and 2.8 to analyze dependence of your alignment on this parameter. Lower pairwise similarity may require somewhat lower gapOpen parameter. A value of 2.4 (gapExtension=0.15) was shown to be optimal for structural similarity recognition with the Gonnet et. al.) matrix, while a value of 2.0 was optimal for the Blosum50) matrix ( Abagyan and Batalov, 1997).
Gap Extension The absolute gap penalty is calculated as a product of gapExtension and the average diagonal element of the residue comparison table.
maxPenalizedGap The maximum penalized gap which is used for Gap Open and Extension
- Click OK and the alignment will be displayed in the alignment editor window at the bottom of the graphical user interface.
- Remember to save the project or write the alignment if you wish to keep the alignment for use at another time.
8.2.6 Sequence to Structure alignment |
This option allows you to align a sequence to a template structure sequence using secondary structure weighting.
- Read into ICM the sequence (ModelSeq) you wish to align to the template sequence.
- Read in the template (TemplateSeq) structure and extract the sequence from this structure
- Bioinfo/Sequence to Structure Alignment
- Enter the ModelSeq and the TemplateSeq name
- Enter the name you wish to call the alignment.
- Enter the weights you wish to use for apha and beta secondary structure. The default values have been very well tested.
- This function uses a dynamic algorithm to find the alignment of the locally structurally similar backbone conformations. The RMSD is calculated within a certain residue window. The default is 3.
- Press OK and the alignement will be displayed in the bottom of the gui interface.
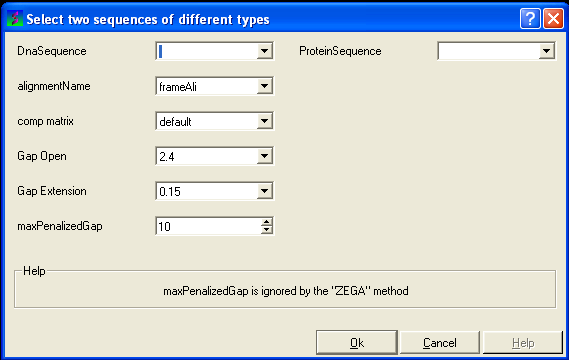
8.2.7 Align DNA vs Protein |
To align DNA to protein:
- Select the 'Bioinfo' menu.
- Select the option Align DNA vs Protein
- Follow the data entry instructions shown in the previous section entitled "align two sequences" but enter one DNA sequence and one protein sequence.
8.2.8 Multiple Sequence Alignment |
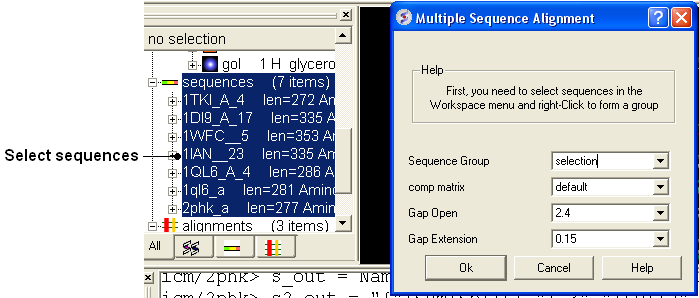
To align more than 2 sequences:
- Read into ICM the sequences you wish to align.
- Select the sequences you wish to align in the ICM workspace. A sequence can be selected by double clicking (highlighted blue in ICM workspace) - a range of sequences in the ICM Worskpace can be selected by holding down the SHIFT button and double clicking. A non-contiguous selection can be made by holding down the CTRL button and double clicking.
- Bioinfo/Multiple Sequence Alignment
- Enter the name of the sequence group. If you selected the sequences as described above then the name of the group is selection. Other named groups of sequences can be made by right clicking on the sequence selection.
- Select the comparison matrix you would like to use.
- Enter Gap open and extension values.
Gap Open The absolute gap penalty is calculated as a product of gapOpen and the average diagonal element of the residue comparison table You may vary gapOpen between 1.8 and 2.8 to analyze dependence of your alignment on this parameter. Lower pairwise similarity may require somewhat lower gapOpen parameter. A value of 2.4 (gapExtension=0.15) was shown to be optimal for structural similarity recognition with the Gonnet et. al.) matrix, while a value of 2.0 was optimal for the Blosum50) matrix ( Abagyan and Batalov, 1997).
Gap Extension The absolute gap penalty is calculated as a product of gapExtension and the average diagonal element of the residue comparison table.
8.2.9 Link to Structure |
To link a structure to an alignment:
- Double click on the structure in the ICM workspace to select it.
- Bioinfo/Link to Structure
| NOTE Links are described in more depth in the Making Links Section of the manual. |
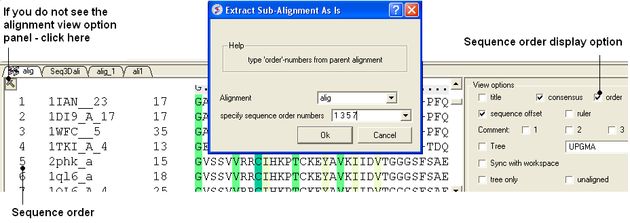
8.2.10 Extract Sub-Alignment As Is |
On occasion you may want to extract a sub alignment from a bigger alignment. For example you wmay only wanto extract the alignment for the sequences linked to a structure.
To extract a sub-alignment:
- An initial multiple sequence alignment must first be displayed in the graphical user interface.
- Bioinfo/Extract Sub-Alignment As Is
- Enter the name of the algienment from which you wish to extract a sub-alignment from.
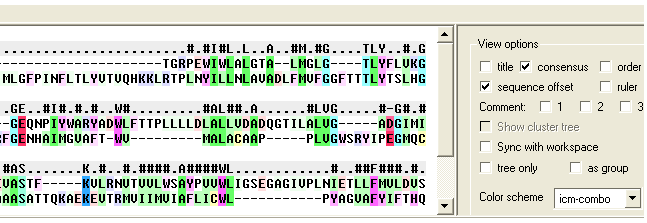
- Specify the sequence order numbers you wish to extract - enter each number separated by a space.You can see the sequence order alignment number by selecting the order option in the alignment view options panel. See image below below.
- Click OK and the extracted sequence alignment will be displayed in a separate alignment tab.
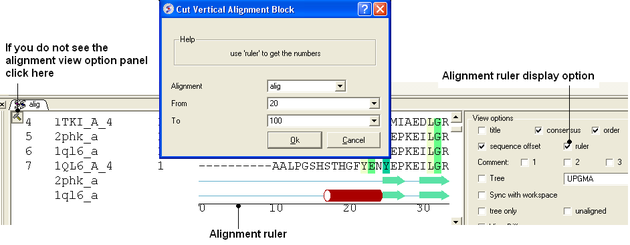
8.2.11 Cut Vertical Alignment Block |
To cut a vertical alignment block:
- An initial alignment must first be displayed in the graphical user interface.
- Bioinfo/Cut Vertical Alignment Block
- Enter the alignment from which you wish to cut from.
- Enter the region of the alignment you wish to cut (from: to:). The easiest way to determine the region to cut is to display the ruler in the alignment. This is an option in the alignemtn view panel - see image below.
- Click OK and the cut section will be displayed in a new alignment.
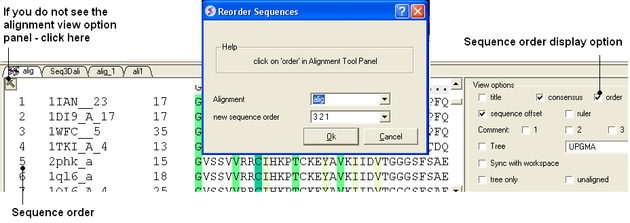
8.2.12 Reorder Sequences |
To reorder sequences in an alignement
- An initial multiple sequence alignment must first be displayed in the graphical user interface.
- Bioinfo/Reorder Sequences
- Enter the alignment name
- Enter the new sequence order.You can see the sequence order alignment number by selecting the order option in the alignment view options panel. See image below below.
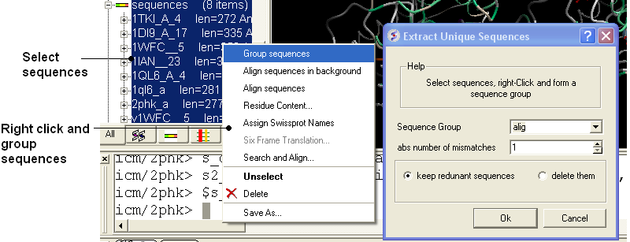
8.2.13 Extract Unique Sequences |
To extract unique sequences from a group of sequences:
- Read into ICM the sequences you wish to make unique.
- Select the sequences. A sequence can be selected by double clicking (highlighted blue in ICM workspace) - a range of sequences in the ICM Worskpace can be selected by holding down the SHIFT button and double clicking. A non-contiguous selection can be made by holding down the CTRL button and double clicking.
- Right click on the sequence selection in the ICM Workspace and select Group sequences
- Bioinfo/Extract Unique Sequences
- Enter the name of the sequence group.
- Enter the number of residue mismatches necesary to determine that a sequence is unique or not.
- Select whether you want to keep the redundant sequences or delete them from ICM.
8.2.14 Load Example Alignment |
To see an example of an alignment select:
- Bioinfo/Load Example Alignment
| Prev From File | Home Up | Next Search and Align |