| Prev | ICM User's Guide 8.4 Sequence Alignments | Next |
[ Introduction | Two Seqs | DNAtoProtein | Multiple | Drag & Drop ]
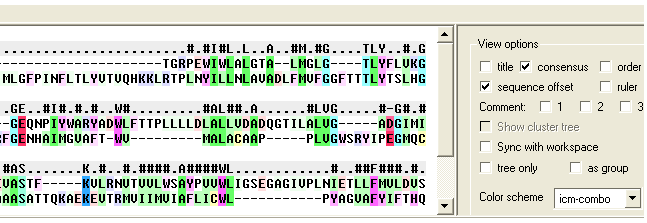
ICM provides a powerful sequence alignment editing tool.
You can customize your sequence alignments in a number of ways:
- Coloring according to a number of different consensus schemes.
- Customizing your own consensus tables.
- Shading areas of interest.
- Boxing areas of interest.
- Adding comments to an alignment.
- Saving an alignment as a high quality image for publication.
- Displaying and analyzing phylogenetic trees.
- Direct selection from the alignment to the 3D object.

8.4.1 Alignment Introduction |
To align two or more sequences you need to use the options in the 'Bioinfo' menu shown below.
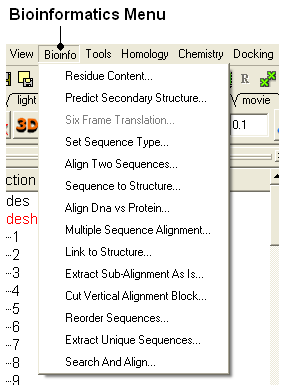
To construct an alignment, two or more sequences need to be loaded into ICM. This can be done be done in one of the following ways:
- Constructing your own sequence see new sequence section.
- Extracting the sequence from a loaded PDB sequence by:
- File/Open sequence in FASTA seq format.
- File/Load SwissProt
- Right clicking on the object name in the workspace panel
- Select 'extract sequence' and the name of the extracted sequence will be displayed in the terminal window.

Once the alignment has been constructed it will be displayed at the bottom of the graphical user interface (see below). If you cannot see the alignment try the Windows menu and select alignments.

8.4.2 Align Two Sequences |
To align two sequences:
- Select the 'Bioinfo' menu.
- Click on 'Align Two Sequences' and the following data entry box will be displayed.
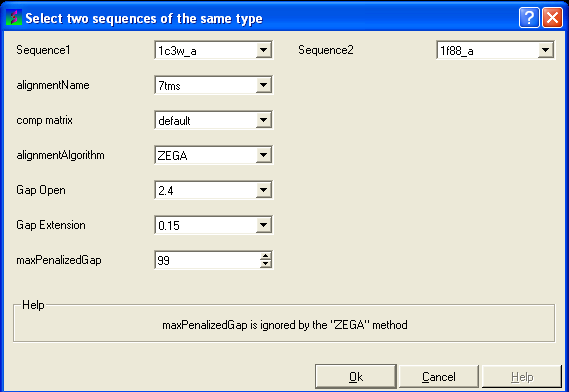
- Enter the name of your first sequence in the 'Sequence 1' data entry box.
- Enter the name of your second sequence in the 'Sequence 2' data entry box.
| NOTE: Any sequences already loaded into ICM can be seen by clicking on the down arrow next to the 'Sequence 1 and 2' data entry boxes. This can save typing and trying to remember what you called your sequence. |
- Enter a unique alignment name in the 'alignmentName' data entry box.
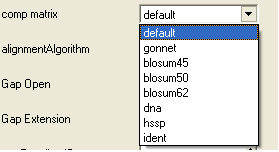
- Select a comparison matrix from the list shown below by clicking on the arrow next to the 'comp matrix' data entry box.
- Select the alignment algorithm you wish to use from the list shown below by clicking on the arrow next to the 'alignmentAlgorithm' data aentry box.
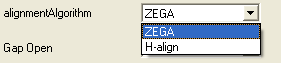
ZEGA - a Zero End-gap Global Alignment, that is a pairwise alignment method based on the Needleman and Wunsch algorithm modified to use zero gap end penalties. This type of alignment was first described by Michael Waterman, who called it the "fit" alignment. The paper of Abagyan and Batalov, 1997 describes the statistics of the structural significance of the alignment score and optimization of the alignment parameters for the best recognition of structurally related proteins.
H-Align - alignment method used in the Align and Score functions and find database command (as described in Batalov and Abagyan, 1999)
- Enter the values you wish to use for Gap Open, Gap Extension and the maximum penalized gap penalty.
Gap Open The absolute gap penalty is calculated as a product of gapOpen and the average diagonal element of the residue comparison table You may vary gapOpen between 1.8 and 2.8 to analyze dependence of your alignment on this parameter. Lower pairwise similarity may require somewhat lower gapOpen parameter. A value of 2.4 (gapExtension=0.15) was shown to be optimal for structural similarity recognition with the Gonnet et. al.) matrix, while a value of 2.0 was optimal for the Blosum50) matrix ( Abagyan and Batalov, 1997).
Gap Extension The absolute gap penalty is calculated as a product of gapExtension and the average diagonal element of the residue comparison table.
maxPenalizedGap The maximum penalized gap which is used for Gap Open and Extension
- Click OK and the alignment will be displayed in the alignment editor window at the bottom of the graphical user interface.
- Remember to save the project or write the alignment if you wish to keep the alignment for use at another time.
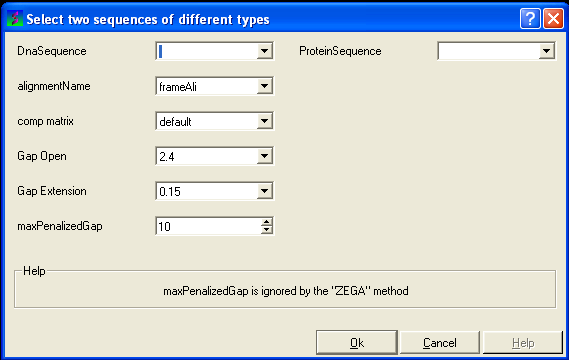
8.4.3 Align DNA to Protein |
To align DNA to protein:
- Select the 'Bioinfo' menu.
- Select the option Align DNA vs Protein
- Follow the data entry instructions shown in the previous section entitled "align two sequences" but enter one DNA sequence and one protein sequence.
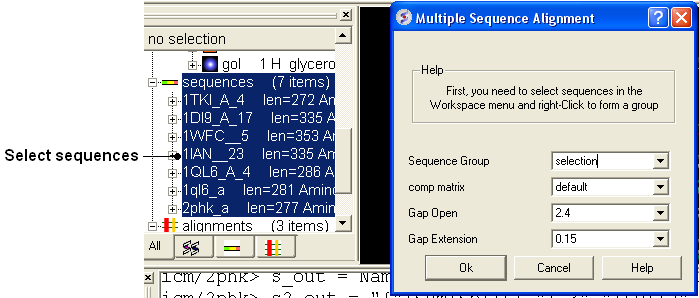
8.4.4 Align Multiple Sequences |
- Read into ICM the sequences you wish to align.
- Select the sequences you wish to align in the ICM workspace. A sequence can be selected by double clicking (highlighted blue in ICM workspace) - a range of sequences in the ICM Worskpace can be selected by holding down the SHIFT button and double clicking. A non-contiguous selection can be made by holding down the CTRL button and double clicking.
- Bioinfo/Multiple Sequence Alignment
- Enter the name of the sequence group. If you selected the sequences as described above then the name of the group is selection. Other named groups of sequences can be made by right clicking on the sequence selection.
- Select the comparison matrix you would like to use.
- Enter Gap open and extension values.
Gap Open The absolute gap penalty is calculated as a product of gapOpen and the average diagonal element of the residue comparison table You may vary gapOpen between 1.8 and 2.8 to analyze dependence of your alignment on this parameter. Lower pairwise similarity may require somewhat lower gapOpen parameter. A value of 2.4 (gapExtension=0.15) was shown to be optimal for structural similarity recognition with the Gonnet et. al.) matrix, while a value of 2.0 was optimal for the Blosum50) matrix ( Abagyan and Batalov, 1997).
Gap Extension The absolute gap penalty is calculated as a product of gapExtension and the average diagonal element of the residue comparison table.
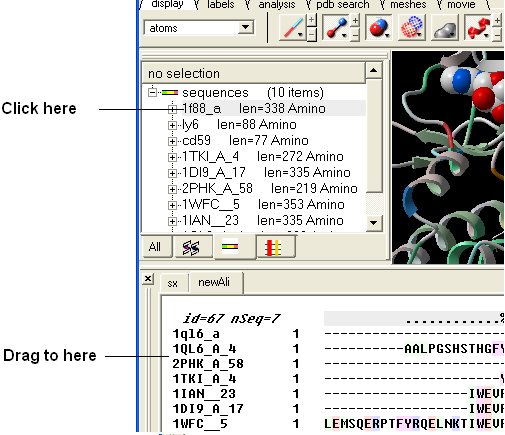
8.4.5 Drag and Drop |
An easy way to add another sequence to an alignment is to drag and drop a loaded sequence from the ICM workspace panel to the alignment window. The sequence automatically becomes part of the alignment.
| Prev Search and Align | Home Up | Next Editor |