| Prev | 4.1 Searching the PDB | Next |
[ PDB | Query | PDB Search Results Table | Patern | Sensitive | Results | Load | Header ]
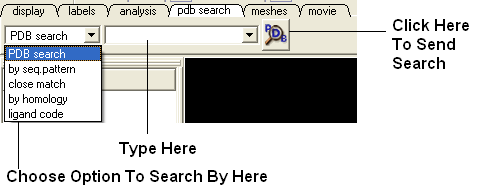
The PDB search tab provides easy access to the PDB database. You can use keyword searching or type in the PDB code you are interested in. An asterisk (*) wildcard can be used to list all the pdb files currently available in the protein databank. Different fields can be searched by using the drop down arrow as shown below. More advanced PDB search tools and how to use the PDB search result table are described in the section entititled Searching the PDB.
Once a search is complete a table of PDB files relating to your search query will be displayed. To view the PDB file in 3D in the graphical display double click on a row in the PDBSearchResults table.
| NOTE: If you have a PDB structure already saved you can read it into ICM by going to the File Menu and selecting Open. PDB files that have been viewed previously can be loaded using File/ Recent PDB Codes. |
4.1.1 Searching the PDB |
Protein structures solved by X-ray crystallography, NMR or other experimental methods are stored in the Protein Data Bank (PDB). These structures can be easily accessed, displayed and analyzed using ICM.
There are four ways to find a structure from the PDB database and load it into ICM:
- Query by keyword or PDB code.
- Query by sequence pattern.
- 2Query by sensitive similarity search.
- Load the PDB file directly from FTP, http, local drive.
4.1.2 Querying PDB by Keyword or PDB Code |
There are four ways of querying the PDB using ICM and keywords. OPTION 3 allows for a much more refined search.
OPTION 1:
- Select Edit/PDB search and the "Find PDB Entries by Keyword" data entry window will be displayed.
- Enter a keyword or PDB code into the Keywords data entry field.
| NOTE: If a keyword has been entered previously it will be available by clicking on the drop-down button. |
- Click the OK button and a list of related PDB entries will be displayed in the PDBSearchResults table of the graphical userinterface.
OPTION 2:
Use the pdb search tab on the tool bar. Select which parameter you wish to search by. Enter some text and this will be searched against the PDB.
Seq Pattern- Enter a protein sequence and this option will tell you whether a protein structure exists in the PDB for that sequence.
Close Match - Enter a protein sequence and this option will tell you which sequences are similar to your entered sequence.
Homology - Enter a protein sequence and homolopgous proteins in the PDB will be displayed in a table.
Ligand Code - Enter the PDB ligand code.(e.g. 1crn)

- Click the OK button and a list of related PDB entries will be displayed in the PDBSearchResults table of the graphical userinterface.
OPTION 3:
- Select Edit/PDB search by field and the ** Find PDB Entries by Keywords and Fields ** data entry window will be displayed.
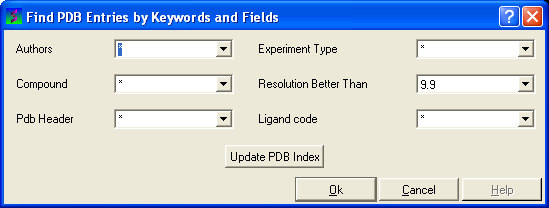
- Enter your search words into the appropriate fields: Author; Compound; PDB Header; Experiment Type; Resolution Better Than
- Click the OK button and a list of related PDB entries will be displayed in the PDBSearchResults table of the graphical userinterface.
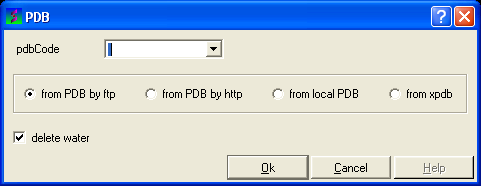
OPTION 4:
- Select File/Load and the PDB data entry window will be displayed.
- Enter the PDB code and select the source of your PDB file.
- Click the OK button and a list of related PDB entries will be displayed in the PDBSearchResults table of the graphical userinterface.
4.1.3 PDB Search Results Table |
- To load a pdb file double click on the search results table.
- Sort the table by right clicking on the column header. Other table manipulation options are described in the Working with Tables chapter.

4.1.4 Querying PDB by Sequence Pattern |
There are two ways to query the PDB by sequence pattern.
OPTION 1:
- Select Edit/PDB Search by sequence pattern
- Cut and paste or type your sequence into the Sequence data entry field.
- Choose whether you wish to display All entries or Entries with unique sequence by checking the appropriate button.
- Click the OK button.
OPTION 2:
Use the word search option on the tool bar

4.1.5 Sensitive PDB Similarity Searches |
There are two ways to search a sequence against the PDB database.
OPTION 1:
If your sequence is already loaded into ICM:
- Select Edit/PDB Search by sensitive similarity
- Type the sequence name into the Sequence name field. Sequences which are already loaded into ICM can be seen by clicking the drop-down button
- Select the number of hits you wish to see by typing the number into the Limit field. A number can also be selected by clicking on the up and down arrows. (Default is 50)
- Select the sensitivity of your search by typing a value into the Expect field. This value is a database-size error estimate and the default value is 0.01.
- Choose whether you wish to display All entries or Entries with unique sequence by checking the appropriate button.
- If you wish to load the sequences leave the Load Sequences box checked.
- If you merely want to see the PDB codes which are similar to your sequence then un-check the Load Sequences box.
- Click the OK button.
OPTION 2:
If your PDB sequence is not loaded into ICM:
- Select Edit/Search with external sequence
- Cut and paste or type (shown below) your sequence into the Sequence data entry field.
- Select the number of hits you wish to see by typing the number into the Limit field. A number can also be selected by clicking on the up and down arrows. (Default is 50)
- Select the sensitivity of your search by typing a value into the Expect field. This value is a database-size error estimate and the default value is 0.01.
- Choose whether you wish to display All entries or ** Entries with unique sequence ** by checking the appropriate button.
- If you wish to load the sequences leave the Load Sequences box checked.
| NOTE: If you merely want to see the PDB codes which are similar to your sequence then un-check the Load Sequences box. |
- Click the OK button.
| NOTE: You can also use the toolbar search option by homology if you wish. |
4.1.6 Working with PDB Search Results |
Once you have searched for a PDB structure, a table with the search results will be displayed on the bottom of the ICM window. See the Tables section for more information on how to use ICM tables. See the next section loading your PDB file for information how to view the PDB file. More information about working with tables can be found in the Tables Section of this manual.
4.1.7 Loading Your PDB File |
- To load a pdb file double click on the search results table.
- Sort the table by right clicking on the column header. Other table manipulatio n options are described in the Working with Tables chapter.

4.1.8 Display PDB Header |
To display the PDB Header for a PDB file.
- First load a PDB file into ICM (see Load PDB)
- Double click on the word header in the ICM Workspace.

- The PDB Header information will be displayed.
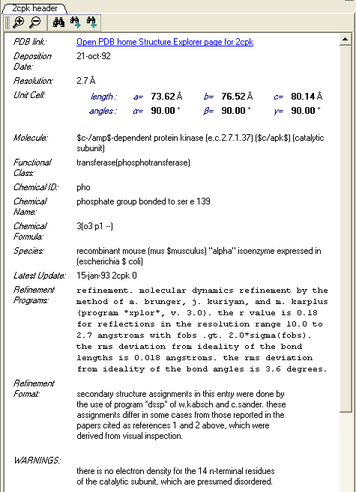
- Click on the blue hyperlinked text to link to external web pages for additional information if needed.
| Prev Protein Structure | Home Up | Next Convert |