Google Search: Keyword Search:
| Prev | ICM User's Guide 10.28 Cluster Set | Next |
[ Chemical clustering | Center | Re-order and Distance | Edit tree | Auto Close | Max Common Substructure | R-Group Decomposition ]
| Available in the following product(s): ICM-Chemist | ICM-Chemist-Pro | ICM-VLS |
Clustering is described in more detail in the Tables Clustering section of this manual. To undertake chemical clustering choose:
- Chemistry/Cluster Set
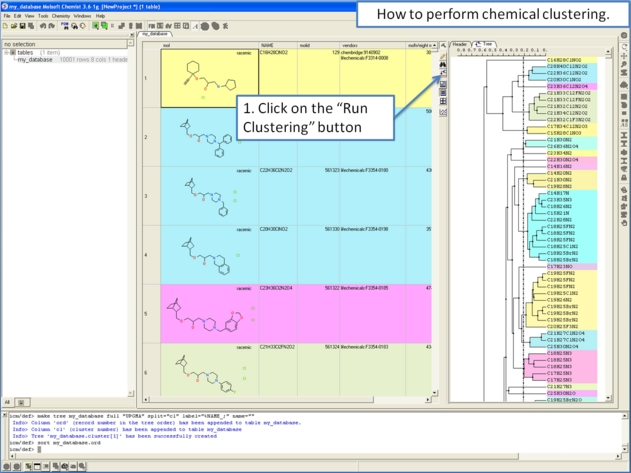
10.28.1 How to perform chemical clustering. |
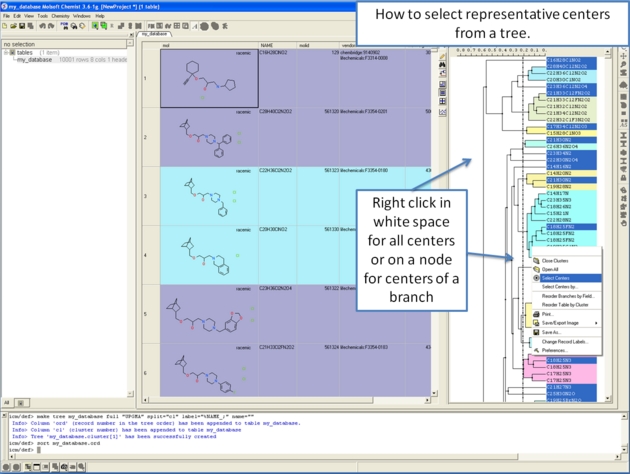
10.28.2 How to select representative centers from a tree. |
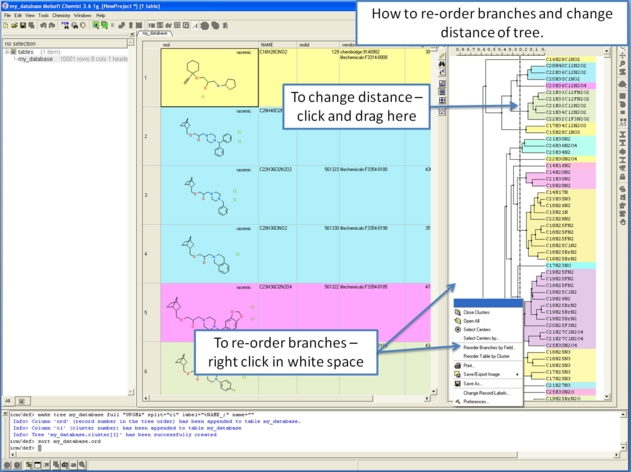
10.28.3 How to reorder branches and change the distance of trees. |
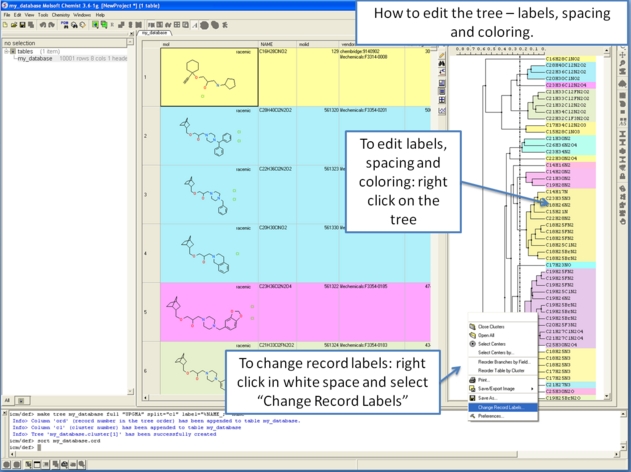
10.28.4 How to edit the tree - labels, spacing and coloring. |
10.28.5 Auto Close Tree Mode |
To make working with large cluster trees a little easier you can activate the auto close mode which will close downstream clusters and make them more compact.
To do this:
- Right click in white space on the cluster and choose Preferences.
- Select "Auto Close Clusters Downstream".
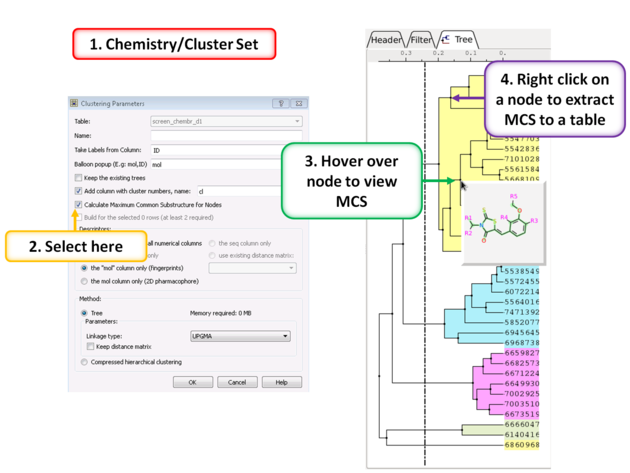
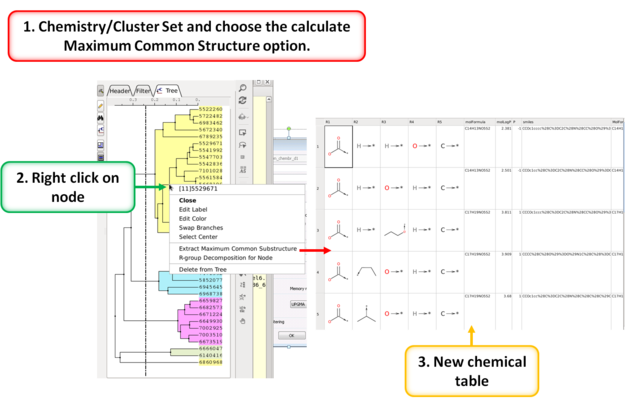
10.28.6 Calculate Maximum Common Substructure for Nodes |
To calculate the Maximum Common Substructure (MCS) for nodes in a tree:
- Chemistry/Cluster Set
- Select the option Calculate Maximum Common Substructure for Nodes
- Hover over a node to view the MCS.
- Right click on a node and choose "Extract Maximum Common Substructure" to extract it to a table.
10.28.7 R Group Decomposition from Clustering Node |
To perform R-group decomposition directly from a cluster node:
- Chemistry/Cluster Set
- Select the option Calculate Maximum Common Substructure for Nodes.
- Hover over a node to view the MCS.
- Right click on a node and choose "R-group decomposition for node".
The resulting table can be used for R-Group Enumeration or SAR.
| Prev Strain | Home Up | Next PCA Analysis |