| Prev | ICM User's Guide 12.3 Build Model | Next |
To build a molecular model:
- Click on the 'Homology' menu at the top of the graphical user interface.
- Select Build Model and a data entry box will be displayed as shown below.
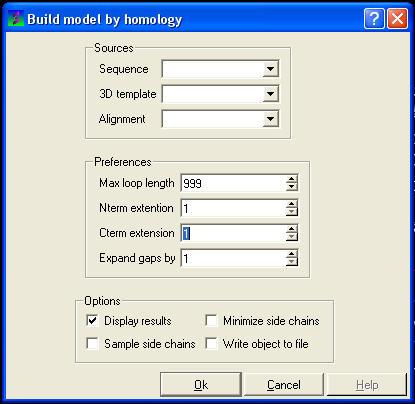
This data entry box is split into 3 sections, the first is 'sources' where you need to specify your query sequence, template and alignment. The second section is called 'preferences where penalty information for the model needs to be entered and the third section is called 'Options'. Each will be described in detail below.
To construct your model follow these steps:
- Enter the name of your QUERY (ie the sequence of the model you wish to build - NOT the template) sequence in the data entry box labeled 'sequence. If you click on the arrow next to this box a list of sequences loaded in ICM will be displayed click on your QUERY sequence. The names of the sequences are also listed in the workspace panel on the left of the graphical user interface.
- Enter the name of your template structure in the '3D template' data entry box. Once again the name of your template structure can be found by clicking the down arrow or in the workspace panel.
- Enter the name of your alignment in the 'Alignment' data entry box.
You could build your model now as ICM has enough infromation but it may be wise to take a look at some of the preferences and change them accordingly. However in most cases the default values provided are sufficient to produce a good quality model.
To change the preferences either type the number you wish or use the up and down arrows next to the data entry boxes.
Max loop length (default= 999) - loops longer than this value are not modeled
Nterm extension (default=1) - the maximal length of the N-terminal model sequence which extends beyond the template
Cterm extension (default=1) - the maximal length of the C-terminal model sequence which extends beyond the template
Expand gaps by (default=1) - additional widening of the gaps in the alignment. End gaps are not expanded
Now all you need to do to build your model is to select some options. Check the box if you would like ICM to perform that option.
The options are:
Display results - displays your model in the 3D graphics window
Minimize side chains - performs minimization on the side-chains
Sample side chains - performs monte-carlo optimization on the side chains
Write object to file - writes your new model as an ICM object
To build your model:
- Click OK
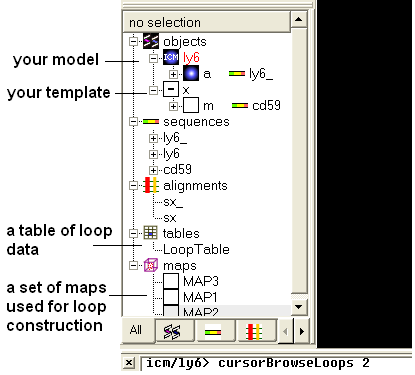
Once your model is built a new object will be seen in your workspace panel. This is your model (see below).
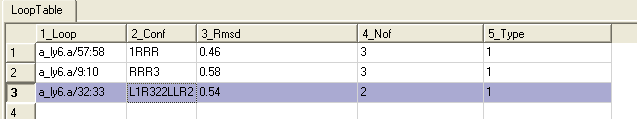
A table of the loop data will also be displayed showing the RMSD from the template.
| Prev Start | Home Up | Next Interactive Modeling |