| Prev | ICM User's Guide 12.2 Getting Started | Next |
The three items you need for ICM protein molecular modeling are:
- An alignment (see alignment section) of your query sequence against a template sequence from the PDB. This is a *.ali file in ICM.
- A template structure from the PDB converted (See convert object) into an ICM object.
- A sequence file of your query sequence for the structure you wish to allographic-construct and a sequence file for your template. Note ICM automatically extracts this information from the alignment or template structure.
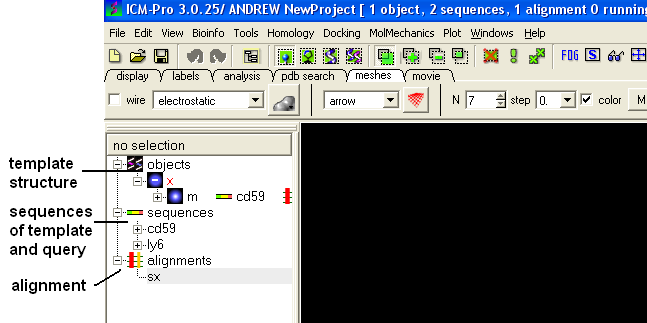
Your graphical user interface window should look something like this:
| Prev Introduction | Home Up | Next Build |