| Prev | ICM User's Guide 12.4 Interactive Modeling | Next |
[ Make | Modeler's View | Interactive Loop ]
How to perform interactive modeling:
What you need before you can undertake interactive modeling:
- A template structure from the PDB converted (See convert object) into an ICM object.
- A sequence file of your query sequence.
You can use an alignment you have constructed yourself or allow ICM to generate one (referred to as automatic)
- An alignment (see alignment section) of your query sequence against the template structure from the PDB. This is a *.ali file in ICM.
12.4.1 Making an interactive model. |
- Homology/Interactive Modeling
- Enter the name of your loaded sequence.
- Enter the name of your loaded 3D template
- Enter the name of your alignment or allow ICM to generate an alignment by selecting automatic
- Check whether you wish ICM to sample the loop regions of your model or not
An Interactive Modeler's View of the alignment will then be displayed.
12.4.2 Modeler's View |
Once you have made your interactive model your graphical user interface should look something like this:
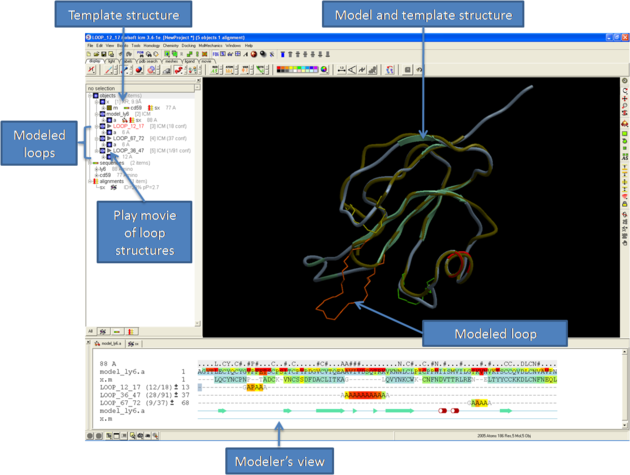
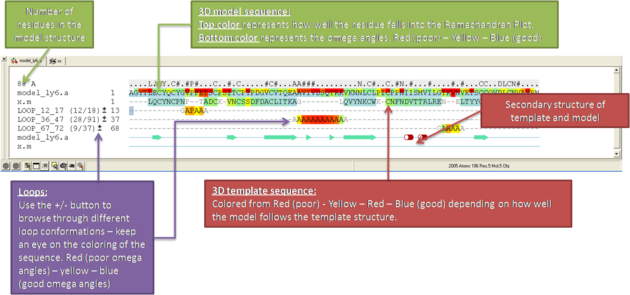
What do all the elements of the Modeler's View mean?
12.4.3 Interactive Loop Modeling |
You can browse a number of different loop conformations by clicking on the +/- button next to the loop in the modeler view's window. Keep an eye on the colors of the loop residues in the alignment - they are colored red (poor) -yellow - blue (good) depending on how good the omega angles are. The residues in the loops are represented by Alanines for every residue other than Glycine or Proline.
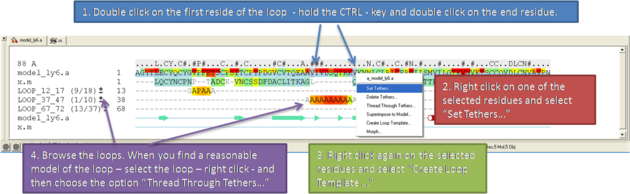
If you want to remodel the loop.
- Set tethers at the residue at the start and end of the loop. To set a tether double click on the two residues whilst holding down the CTRL key. Right click and select Set tethers. Selecthow you want to tether to the template either by alignment, residue numbering or selection. Select the alignment - automatic represents the Moldeler's View. Select the Template Molecule which is the object you built the model on.
- Select the loop you wish to model including the tethered residues at the start and end of the loop and then right click and select Create Loop Template
- Once the simulation has finised you can once again browse the solutions by clicking on the +/- button next to the loop in the modeler view's window.
- When you have identified a reasonable conformation of the loop you can re-thread it onto the model structure by selecting the loop region in the Modeler's View. You can do this by clicking and dragging over the loop. Right click and select Thread Through Tethers.
| Prev Build | Home Up | Next Display Loops |