| Prev | ICM User's Guide 12.6 Loop Modeling | Next |
Building an accurate model of a loop is very tough. However with small loops ICM has been very succesful. ICM was used to design two new 7 residue loops and in both cases the designs were successful. Moreover, the predicted conformations turned out to be exactly right (accuracy of 0.5�) after the crystallographic structures of the designed proteins were determined in Rik Wierenga's lab.
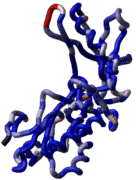
To build a new loop to an existing structure or to improve your already modeled loops:
- Read your modeled structure into ICM. Or continue immediately after using build model.
- Select the loop region you wish to model (green crosses in the graphical display).
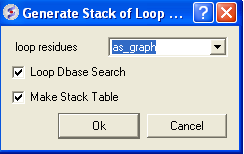
- MolMechanics/Sample Loop.
- MolMechanics/View Stack A table will then be displayed with the optimized structures ranked by energy. To view each structure double click on the table. The first row is the loop with the best energy.
| Prev Display Loops | Home Up | Next Regul |