| Prev | ICM User's Guide 18.7 Find Best Replacement Group | Next |
Find best replacement group for selected atom
- Select the atom you want to add a new replacement group to.
- Click on the Advanced/Find Group button.
- A dialog box as shown below will be displayed. Select whether you want the substituent score only to be evaluated (quick) or the full binding score.
- Select whether you want to screen the modifying groups built into ICM (see sarray of smiles called LIGAND.modifiers) or a table of your own modifier groups. If you choose your own table you will need to load the table (sdf file) into ICM and enter the name of the table into this dialog box or you can add modifiers to the sarray of smiles called LIGAND.userModifiers.
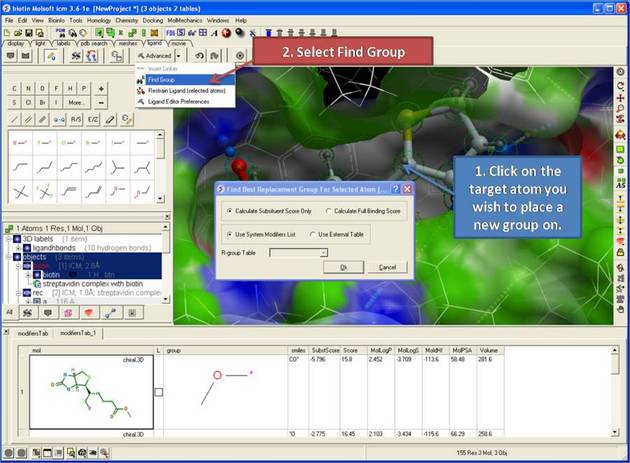
- ICM will add each fragment to the target atom and sample the energy and return a table ranked by score (see below).
| Prev Ligand linker | Home Up | Next Ligand Tethers |