| Prev | ICM User's Guide 3.3 How to Work With Sequences and Alignments | Next |
[ SwissProt | FASTA | New Sequence | Extract Sequence | Sequence Alignment | Drag & Drop ]
A quick start guide on how to search for, read in, analyze sequences and build alignments.
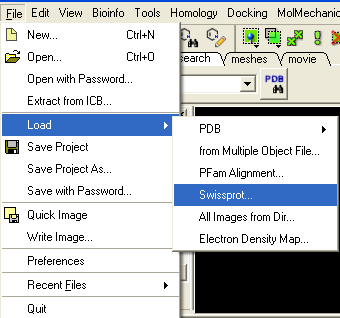
3.3.1 How to Download a SwissProt sequence |
To download a SWISSPROT sequence into ICM
- Select File/Load/SwissProt
- The sequence will be loaded into the ICM Workspace.
3.3.2 How to Load a FASTA Format File |
To read a FASTA file:
- Select File/Open and look for file type "Sequence Format"
- The sequence will be loaded into the ICM Workspace.
3.3.3 How to Make a New Sequence |
- File/New
- Select "sequence" tab.
- Cut and Paste or type sequence
- Click OK and the sequence will be loaded into the ICM Workspace.
3.3.4 How to Extract a Sequence from a PDB Structure. |
- As an example we will use the PDB structure 1STP. Type 1STP in the pdb search tab and press return.
- Right click on the protein molecule "m" and select "Extract Sequences"
- The sequence will be loaded into the ICM Workspace.
3.3.5 How to Make a Sequence Alignment |
An example of how to perform an alignment between two sequences.
PDB Search:
- PDB Search Tab 1ql6
- PDB Search Tab 1ian
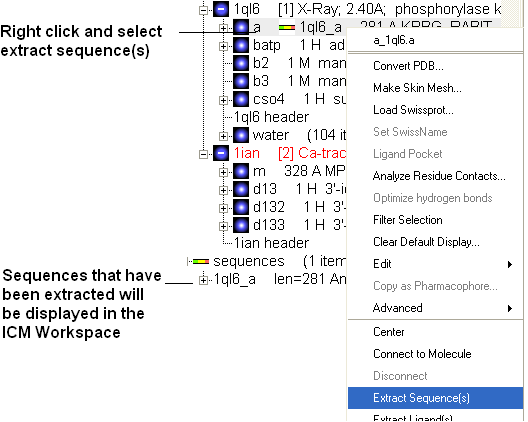
- Now extract the sequence information from each protein. To do this right click on the molecule "a" of 1ql6 and molecule "m" of 1ian. and select extract sequences. Once the sequences have been extracted you should see the sequence in the ICM Workspace entitled 1ql6_a and 1ian_m
- Now align the sequences by selecting both sequences right clicking and selecting Align sequences. An alignment will be displayed at the bottom of the graphical user interface.
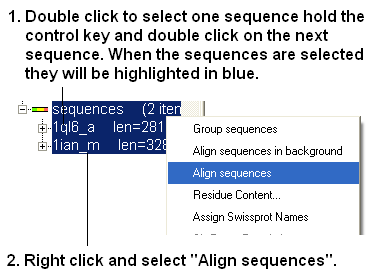
| NOTE: To build a multiple alignment just select more sequences right click and align sequences. Or you can drag and drop sequences into an already made alignment. |
3.3.6 How to Make an Alignment using Drag and Drop |
(An easier way to build an alignment is to drop one sequence onto another in the ICM Workspace}
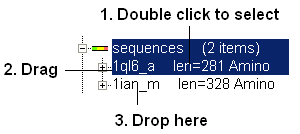
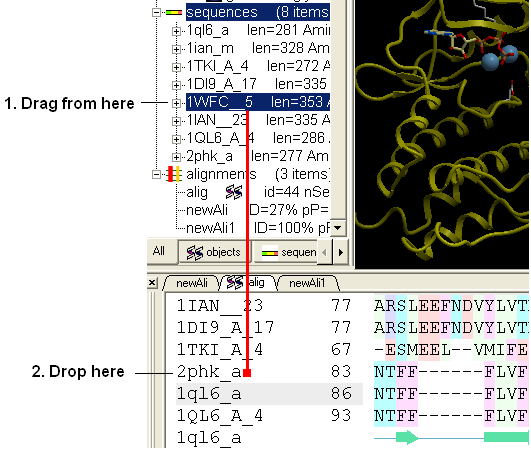
You can add sequences to an alignment by dragging and dropping them into the alignment.
| Prev Links | Home Up | Next Protein Structure |