| Prev | ICM User's Guide 11.1 Crystallographic Neighbor | Next |
Theory
Molecular objects and 3D density maps may contain information about crystallographic symmetry. It consists of the following parameters:
- Crystallographic group eg. P2121 that determine N (depends on a group) transformations for the atoms in the asymetric unit.
- Crystallographic cell parameters A, B, C, Alpha, Beta and Gamma
To generate the coordinates within one cell one needs to apply N transformations and then to generate neigboring cells the content of one cell needs to be translated in space according to the cell position.
ICM has a function which generates crystallographic neighbors for the selected atoms. For large proteins it is impractical to generate neighbors for the whole molecule due to the high number of atoms in all neighboring molecules.
This information allows to generate symmetry related parts of the density or molecular objects.
To generate symmetry related molecules around a selection of atoms:
- Read a PDB file into ICM. For instruction see the section entitled Finding a PDB Structure.
- Display the structure and select the residues around which the symmetry will be generated. For information on how to select residues see the Making Graphical Selections section.
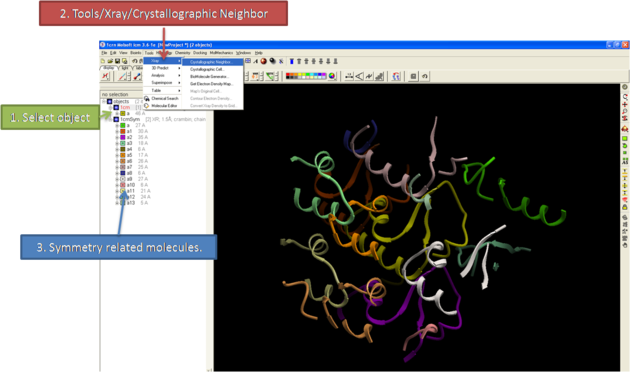
- Select the menu Tools/Xray/Crystallograhic Neighbors.
A data entry box as shown below will be displayed.
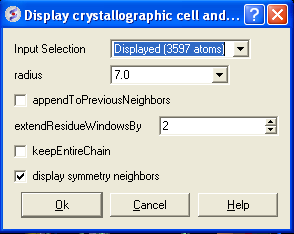
- Select the object.
- Enter the radius around your selction from which you wish to construct the symmetry related molecules.
- If you have made symmetry related molecules previously you can select appendToPreviousNeighbors otherwise leave unchecked.
- The extendResidueWindowsBy option will allow a window of residues outside of the selection radius selected above to be displayed
- If you leave the keepEntireChain unchecked then a fragment of each neighbor will be created. If you check this box the full neighbor will be generated
- Check display symmmetry neighbors to display them in the graphics window. The nearest neighbor residues will be displayed in xstick representation and the each neighbor colored by molecule.
- Click OK.
The crystallographic symmetry neighbors will be displayed in the Workspace. By default the object will have the object name + "Sym" and each of the neighbors will be individual molecules.
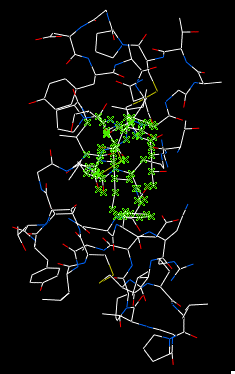
For packing analysis and display you can color each symmetry unit a different color as described in the Structural Representations Color section. This is shown in the picture below.
| Prev Crystallographic Analysis | Home Up | Next Crystallographic Cell |