| Prev | ICM User's Guide 21.6 FAQ-Cheminformatics | Next |
[ MolCart License | Connect to MolCart | MolCart Compounds | MolCart SDF | Chemical Search | Molcart New Database | Molecular Editor | convert to 3D | Grid View | Convert Chemical | convert to 3D and preserve | Add Property Column | Extract Ligand | Druglikeness | Chemical Monitor | Faq molcart text search | SMILES | APF Model | Chemical Rotate ]
Frequently asked questions regarding small molecules, ICM-Chemistry tools and MolCart
- How do I generate the hostid for my MolCart license?
- How do I connect to Molcart?
- How can I download the MolCart vendor compounds provided by MolSoft?
- I have a database in MolCart and I want to save it in SDF format - how can I do this?
- How do I perform a chemical search?
- How do I make a new molcart database from a query search?
- How can I draw small molecules?
- How do I read in a small molecule from ISIS draw and convert it to 3D?
- How can I change the layout of a chemical table?
- How can I convert a chemical in a chemical table into 3D?
- I have a small molecule which already has the 3D coordinates defined. How can I load the molecule and not optimize it so as to preserve the assigned 3D coordinates?
- I have a chemical table displayed - how can I add columns of chemical properties associated with each chemical in my table?
- I have a small molecule displayed in 3D in a loaded PDB file. How can I extract this molecule into an ICM Chemical Table?
- What is considered a good druglikeness value?
- I do not see the chemical property monitor in the molecular editor. Where is it?
- How do I perform a text query on a database in MolCart?
- How do I convert SMILES string into a 2D structure
- Is there a way to build a classification model using the APF output?
- How to rotate a 2D chemical sketch so it fits nicely in its cell in a chemical table?
21.6.1 How do I generate the hostid for my MolCart license? |
- Download MolCart from www.molsoft.com/support and unpack it.
- run run /usr/molcart-1.9-5/sysid and send the number to support@molsoft.com
21.6.2 How do I connect to Molcart? |
When you unpack the MolCart distribution from www.molsoft.com/support you will be given a unique number (which you need to send to support@molsoft.com to get a MolCart license) along with MolCart login details such as Server Name, UserName and Password.
Once you have the MolCart distribution loaded you can connect to MolCart by going to Tools/Chemistry/Connect to Molcart
21.6.3 How can I download the MolCart vendor compounds provided by MolSoft? |
MolCart Compound Database is an up-to-date collection of vendor compound databases. This database is divided into three collections:
- Screening Compounds for cherry picking.
- Building blocks for combinatorial chemistry
Each collection consists of two components:
- a single non-redundant set of compounds, with a list vendors and vendor-IDs for each unique compound
- the original files with the full set of fields as provided by each vendor
The MolCart compounds can be downloaded from http://www.molsoft.com/screening.html then to unpack them type:
zcat vendor.gz | mysql -h<hostname> -u<user> -p<password> molcart_database_name
21.6.4 I have a database in MolCart and I want to save it in SDF format - how can I do this? |
In the terminal window type:
write molcart table="molcart_table_name" "name_of_sdf"
21.6.5 How do I perform a chemical search |
You can search MolCart or Chemical Tables using the ICM Chemical Search Window. This window can be displayed by going to:
- Tools/Chemistry/Chemical Search
OR
- Click on this icon
OR
- Right click on a structure in a chemical table and select Query Molecule.
OR
- Or right click on a database in MolCart you wish to search and select Query...
21.6.6 How do I make a new molcart database from a query search? |
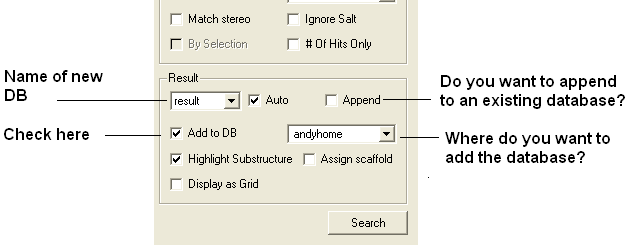
To write the data from a query to a new MolCart database table use the Add to DB option in the Chemical Search window.
21.6.7 How can I draw small molecules? |
Use the molecule editor
Chemistry/Molecular Editor
or look for the ICM molecular editor button at the top of the graphical user interface.
21.6.8 How do I read in a small molecule from ISIS draw and convert it to 3D? |
- Save the molecule in mol format and then read into ICM (File/Open).
- The molecule should be displayed in 2D in a molecular table.
- Right click on the molecule in the table and select chemistry/convert to 3D and optimize.
| NOTE: There is no need to use an external chemistry drawing software when you can use the ICM molecular editor which is fully integrated into the ICM software. |
21.6.9 How can I change the layout of a chemical table? |
To change the layout of a chemical table (eg converting a table to grid view).
- Select the columns you wish to display in grid view. You can do this by clicking on the column headers with the CTRL key pressed down.
- Once the columns are selected right click inside the table and select Table View
- Select Custom Grid...
21.6.10 How can I convert a chemical in a chemical table into 3D? |
To do this:
- Select the chemical or chemicals you wish to convert to 3D. You can do this by clicking on the row number whilst keeping the CTRL key pressed down.
- Right click on the chemical table and select Chemistry/Convert 3D and Optimize
21.6.11 I have a small molecule which already has the 3D coordinates defined. How can I load the molecule and not optimize it so as to preserve the assigned 3D coordinates? |
- File/Open and read in the molecule - it should be then displayed in a molecular table
- Right click on the molecule in the table and select Chemistry/ Load and Preserve Coordinates.
21.6.12 I have a chemical table displayed - how can I add columns of chemical properties associated with each chemical in my table? |
To read a chemical table into ICM:
- File/Open and look for sdf files.
To add a chemical property to the table.
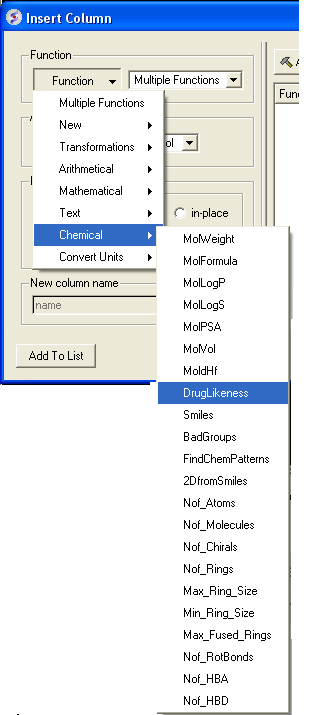
- Right click on the 'mol' column header and select Insert Column...
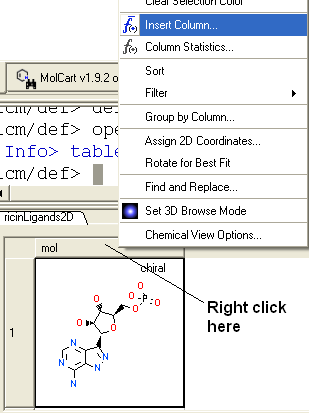
- Click on the drop down 'Function' button and select chemical.
- Select which property you wouls like to add and click OK.
- The property you selected will be displayed in the table.
21.6.13 I have a small molecule displayed in 3D in a loaded PDB file. How can I extract this molecule into an ICM Chemical Table? |
You can extract a ligand from an ICM object or PDB file by:
- Right click on the ligand in the ICM Workspace.=
- Select Extract Ligand.
- Choose to extract either 2D or 3D coordinates and the molecule will be placed in a chemical table.
21.6.14 What is considered a good druglikeness value? |
When building a molecule in the ICM Molecular Editor (Tools/Chemistry) properties such as druglikeness are calculated on the fly. The properties can also be added by inserting a column into a chemical table (right click on column header/ insert column/ Function = Chemical). These values should be used as a guide and druglikeness is a prediction based on drug-like properties. A druglikeness value less than zero indicates that the compound may have some non-drug-like properties.
21.6.15 I do not see the chemical property monitor in the molecular editor. Where is it? |
If you do not see the chemical monitor in the ICM Molecule Editor - Go to:
- Go to Molecular Editor
- View/Chemical Monitor
21.6.16 How do I perform a text query on a database in MolCart? |
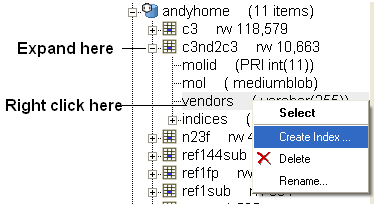
To perform a text search an index needs to be made on the field you wish to search. To do this:
- Expand the contents of the database in the ICM workspace by clicking on the '+' sign.
- Right click on the field you wish to text search.
- Select Create Index
- Select Keyword Search
- Once the index has been created you should see the text query box appear next time you perform a query.
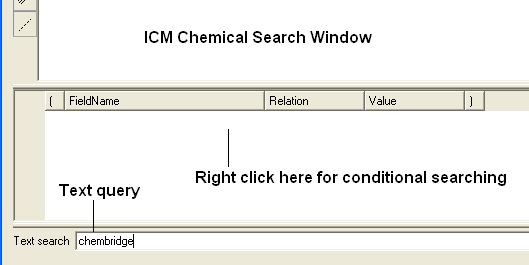
| NOTE: Queries can also be made in the window above the text search window (see above). If you right click you will see fields that you can fill in and use conditional based queries. |
21.6.17 How to convert SMILES strings to 2D |
See How To section.
21.6.18 Is there a way to build a classification model using the APF output? |
_setAPFparams is in the distribution since 3.6-0;
The usage:
icm _apf3Dqsar train=trainingSet.sdf activity=LogIC50 table=testSet.sdf
Training and test set compounds should be all pre-aligned, for example by aligning training set actives using APF multiple chemical alignment, and then superimposing the test compounds onto aligned actives using APF superposition. Any external alignment method can be used as well. The field containing activity data in the training set SDF is specified by activity= argument.
The script also can take alignments in icm multiple object format *.ob, in which case SDFs are only used for input/output of activity data and can be just 2D:
icm _apf3Dqsar train=trainingSet.sdf align=trainingSet3Daligned.ob activity=LogIC50 predict=testSet3Daligned.ob table=testSet.sdf
The results are written to testSet_predict.sdf output file. Some statistics is reported along the way. If testSet.sdf contains activity (i.e. LogIC50) column like the training set, RMSD and R2 will be reported as well.
21.6.19 How to rotate a 2D chemical sketch so it fits nicely in its cell in a chemical table? |
See this description in the command line manual:
http://www.molsoft.com/man/icm-commands.html#make-flat
| Prev Dock Racemic | Home Up | Next FAQ-Simulations |