| Prev | ICM User's Guide 9.3 Contact Areas | Next |
- Read in a protein structure (File/Open or PDB Search)
- Select the region you wish to analyse.
- Tools/Analyze/Contact Areas
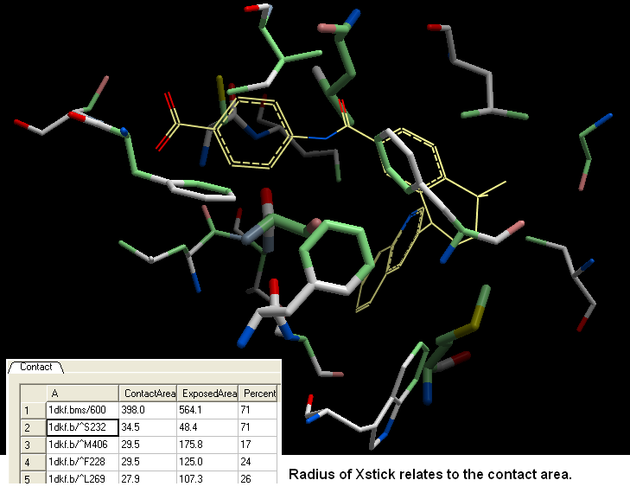
- The xstick display in the region will be scaled according to the atom/residue contact area. For example, residues making large contacts with a ligand will be displayed in thicker xstick representation than those making small contacts.
- A table as shown below will be displayed. Residues making key contacts will be displayed in xstick (radius represents contribution size). Carbon atoms are colored light green, nitrogen atoms are colored light blue and oxygen atoms are colored light red. The table lists the contact area, exposed area and the percentage of contact area compared to exposed.
| NOTE: You can slso right click on the molecule in the ICM Workspace and select "Analyze Residue Contacts" |
| Prev Calculate RMSD | Home Up | Next Closed Cavities |