| Prev | ICM User's Guide 9.1 Find Related Chains | Next |
This option allows you to search the currently loaded PDB files or ICM objects and identify chains which are similar and/or related.
You can do this by:
- Select the objects or pdb files you want to compare.
- Tools/Analysis/Find Related Chains
- Click OK to confirm the selection you made
- A table as shown below will be displayed.
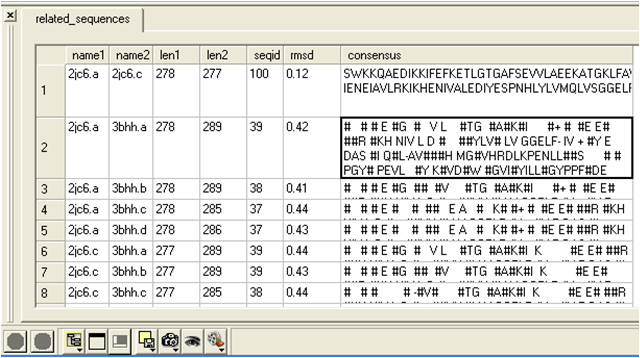
name1 = Name of query structure molecule name2 = Name of hit len1 = length of query len2 = length of hit seqid = Sequence identity percentage consensus = Consensus sequence
| Prev Protein Structure Analysis | Home Up | Next Calculate RMSD |