| Prev | ICM User's Guide 17.6 Protein-Protein Docking | Next |
[ ODA | Setup | P-P Set Project | P-P Receptor Setup | P-P Ligand Setup | Epitope | P-P Maps | P-P Batch | P-P Results ]
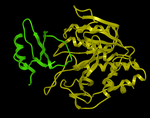
Here we describe the steps for protein-protein docking. An example is described using a complex of subtilisin and chymotrypsin (PDB code:2sni). The example will re-dock the ligand ( PDB code entry 2ci2) into the receptor molecule (PDB code 2st1) and then determine how accurately the molecules are docked by comparison with the complex 2sni. The structure of 2sni is shown below with the ligand displayed in green and the receptor in yellow.
17.6.1 Optimal Docking Area |
The ICM Optimal Docking Area method is a useful way of prediciting likely protein-protein interaction interfaces. If you do not have mutational data or other experimental data which indicates the likely protein-protein docking site this method will be useful. This procedure can save you time during the docking procedure by focusing your docking only on areas on the receptor and ligand most likely to interact.
Theory
ODA (Optimal Docking Areas) is a new method to predict protein-protein interaction sites on protein surfaces. It identifies optimal surface patches with the lowest docking desolvation energy values as calculated by atomic solvation parameters (ASP) derived from octanol/water transfer experiments and adjusted for protein-protein docking. The predictor has been benchmarked on 66 non-homologous unbound structures, and the identified interactions points (top 10 ODA hot-spots) are correctly located in 70% of the cases (80% if we disregard NMR structures).

To display the optimal docking area.
- Convert the PDB file to an ICM object.
- Tools/3D Predict/Protein Interface by ODA
ODA Example with a subtilisin-chymotrypsin complex.
As an example we will determine whether the ICM-ODA method can accurately predict the binding surface of the complex between subtilisin and chymotrypsin. This example is used in the protein-protein docking tutorial below as well.
This complex has been solved experimentally and has PDB id 2sni.
Calculate the ODA for each subunit (Tools/3D Predict / Protein Interface by ODA).
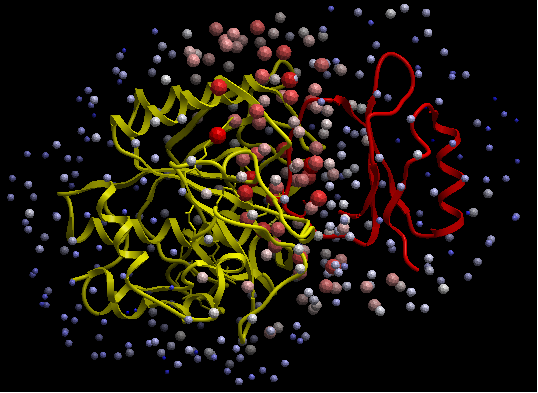
ODA for subtilisin and ODA for chymotrypsin - red colored spheres indicate a region highly likely to be involved in protein-protein interaction, blue coloring is unlikely to be involved in protein-protein interaction. A clickable table is also displayed with ODA values.
17.6.2 Protein-Protein Docking Procedure |
To begin the protein-protein docking procedure:
- Read in the PDB files for 2ci2 (ligand) 2st1 (receptor) and 2sni (complex for comparison). For instructions on how to load a PDB structure into ICM please click here.
- Convert all three PDB files into ICM objects.
- Delete all waters and sulfate ions, you can keep the calcium ions if you wish.
Now go onto the first step of the protein-protein docking protocol which is to Set Project name.
| NOTE: All the protein-proteing docking options can be found in the GUI menu Docking/Protein-Protein. |
17.6.3 Protein-Protein Set Project |
Docking/Protein-protein/Set Project
Start the protein-protein docking project setup by defining the project name:
- Click on Docking/Protein-protein/Set project
- Enter a unique name into the Project name data entry box. Avoid spaces and leading digits in the name. All files related to the docking project will be stored under names, which start from the project name.
Now setup the receptor.
17.6.4 Protein-Protein Receptor Setup |
Docking/Protein-protein/Receptor setup
- Enter the Docking Project name e.g. DOCK1
- Enter the receptor molecule e.g. a_2st1.m (use a_2st1.* if you want to include all molecules such as Calcium ions)
- Click OK
Now setup the ligand.
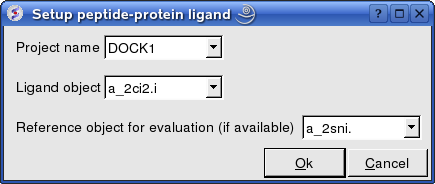
17.6.5 Protein-Protein Ligand Setup |
Docking/Protein-protein/Ligand setup
- Enter the project name e.g. DOCK1
- Enter the ligand molecule e.g. a_2ci2.i
- If you wish to compare your docking data with a solved structure enter the name of the converted reference object in the "Reference Object" data entry box e.g. a_2sni.
- Click OK
Now select an initial point of interest on the receptor referred to as epitope selection (NOTE: This step is optional. If you do not wish to select an initial point of interest junp to the make maps section.
17.6.6 Epitope Selection |
Docking/Protein-protein/Epitope selection
Select an initial point of interest on the receptor for the docking simulation. You may want to check biological data or a reference complex before doing this step.
| NOTE: This step can be left out completely if you dont know or dont want to select an initial point of interest. |
- You can make selections on either the ligand or receptor or both the ligand and receptor. Check the appropriate box(es).
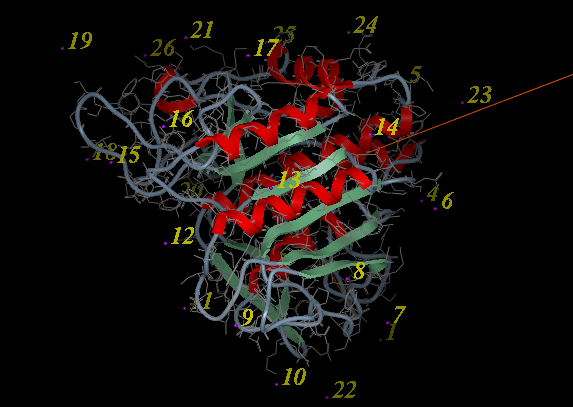
- A display as shown below will be displayed.
- Using the right button of the mouse select a numbered sphere surrounding the receptor or ligand that you wish to dock to by clicking and dragging the mouse over the sphere. The spheres are numbered and change color from purple to yellow when they are selected. If you are happy with the selection type 'a' or press the apply button. The selected numbered regions will change from purple to yellow. The easiest way to select multiple epitopes is to use the pick atom button (green cross button).
- When you have finished selecting the epitoples type 'q' or select the quit button in the terminal window.
NOTE: If you are unsure which epitopes you have selected they are listed in the DOCKING_PROJECT_NAME.tab file in the first two fields
eg here epitope 1 and 3 have been selected in both the ligand and the receptor:
#>I test2.I_selLigPos 1 3 #>I test2.I_selLigRot 1You can also select epitopes by editing this field in the .tab file. |
The next step is to make the maps of the receptor.
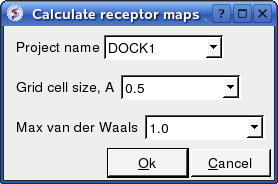
17.6.7 Protein-Protein Make Receptor Maps |
Docking/Protein-protein/Make Receptor Maps
- Enter the Project name e.g. DOCK1
- Enter the grid size e.g. how detailed you want your maps - the default value of 0.5 is generally ok.
- Enter the Max van der Waals value which gives the receptor an element of 'softness' to incoporate some induced fit - the default value of 1.0 is generally ok.
Now run the docking simulation.
17.6.8 Protein-Protein Docking Batch |
The docking can be run on your local machine or in PBS.
To run on your local machine:
Docking/Protein-protein/Docking Batch/Local Machine
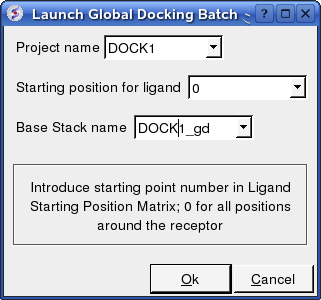
- Enter the Project Name
- Starting Position for Ligand - if you select 0 it will sample all the points on the receptor you selected in the epitope step of the docking project setup. If however you want to break your jobs down into smaller chunks you can enter the number of a position on the receptor you chose in the epitope selection step and it will sample that point.
- Enter a name for the conformational stack which will be saved.
To run in PBS:
Docking/Protein-protein/Docking Batch/PBS
17.6.9 Display Grid Docking Results |
A window will be displayed once the docking has finished or you can check the docking progress by going to Windows/Background Jobs. To display the grid-docking ligand conformations :
- Read object "DOCK1_rec" # read receptor (if not read yet)
- Display a_DOCK1_rec. # display receptor (if not displayed yet)
- Read object "DOCK1_lig" # read ligand object
- Display a_ # display this ligand
- Read table "DOCK1_gd.Var" # read table of the ligand conformations
- Click on table rows to view ligand conformations
- Check the R_Srmsd column for the difference between docked and the crystal structure complex for comparison (if selected).
Or in gui go to:
- Docking/Protein-protein/Docking Batch/Process Global Docking Solutions.
A table as shown below will be displayed. You can sort the table by Energy (ener) by right clicking on the column header and select sort.
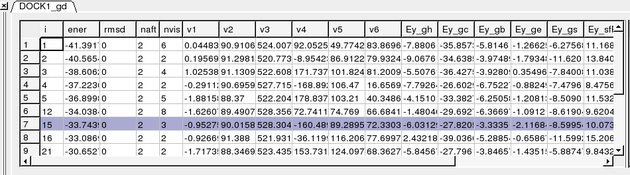
The output columns represent:
- i - a slot number in the stack of conformations
- ener - total energy as calculated before the conformation was stored
- rmsd - the distance (either Cartesian or angular RMSD) between the current conformation of the object and the stack conformation calculated according to the comparecommand.
- naft - the number of visits AFTER the last improvement of energy
- nvis - the total number of visits to this slot; since new conformation are only compared with the last stack conformation the conformations may drift and cover a large area than described by the vicinityparameter
- v1 to v6 - are the virtual variables defining position and rotation of the ligand molecule.
- ey gh - van der Waals grid potential - hydrogen probe
- ey gc - van der Waals grid potential - carbon probe
- ey gb hydrogen bonding grid potential
- ey ge electrobstatics grid potential
- hydrophobic potential
- ey sfPola - polar terms of the solvation energy
- Ey_sfAl - aliphatic terms of the solvation energy
- Ey_sfAr - aromatic terms of the solvation energy
- Ey_compSol - weighted total of the solvation energy terms
To display the complexes:
- Double click on a row of the table shown above. The ligand will be displayed in the ICM workspace and named according to the project name and the conformation number in the stack of solutions (eg DOCK1_lig_conf4 - 4th member of the stack).
- To view interactions between the receptor and the ligand each moleucule needs to be in the same object. See the FAQ section: How do I merge two separate objects into one.
| Prev Autofit | Home Up | Next Small Molecule Docking |