Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 10.20 Convert | Next |
[ 2D to 3D | 3D to 2D Depiction | Convert Smiles to 2D | Convert Structure to Smiles | Convert to Racemic ]
| Available in the following product(s): ICM-Chemist | ICM-Chemist-Pro | ICM-VLS |
10.20.1 Convert 2D to 3D |
- Read a chemical table (sdf file) into ICM.
- Chemistry/Convert/2D to 3D
- Select the table from the drop down list.
- Select to keep hydrogens and/or fix amide bonds.
- Keep current table (In Place) or overwrite.
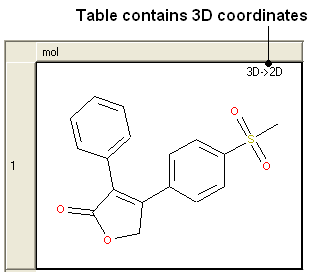
10.20.2 3D to 2D Depiction |
If you have a chemical table displayed containing 3D coordinates or you wish to reassign the 2D cooridnates in an sdf file you can use this option.
- Read a chemical table (sdf file) into ICM.
- Chemistry/Convert/2D Depiction
- Enter name of loaded chemical table.
- Choose In Place if you want to overwrite the table.
- Choose the Files tab to run in batch mode.
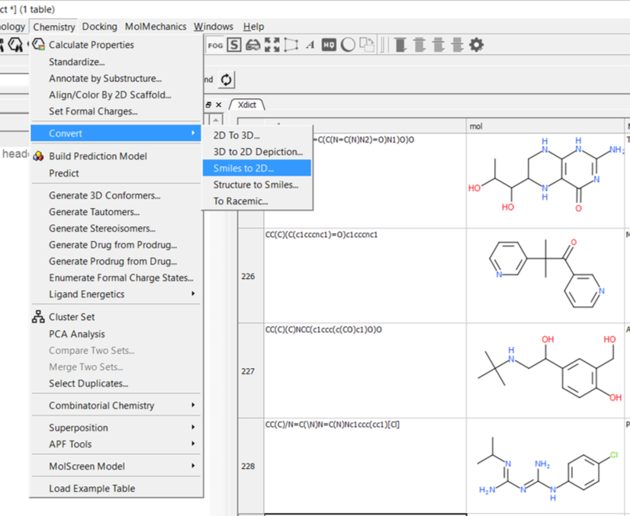
10.20.3 Convert Smiles or InChi to 2D |
To convert smiles or InChi strings to 2D sketches
- Read in a table containing the smiles or InChi strings in separate rows. For example the smiles strings maybe in an Excel file and you can load this into ICM by saving the Excel file as comma-separated (csv).
- Select Chemistry/Convert/Smiles or InChi to 2D.
- Select the table you want to convert using the drop down arrow and the name of the column containing the smiles string.
- Select whether you wish to keep the smiles column in the new table.
- Click OK and a table will be displayed containing the 2D structure.
10.20.4 Convert Structure to Smiles |
To convert an sdf file of 2D or 3D chemical coordinate in Smiles:
- Read a chemical table (sdf file) into ICM.
- Select Chemistry/Convert Structure to Smiles.
- Select the table you want to convert using the drop down arrow and the name of the column containing the 2D sketch.
- Select whether you wish to keep the 2D sketch column in the new table.
- Click OK and a table will be displayed containing the smiles string.
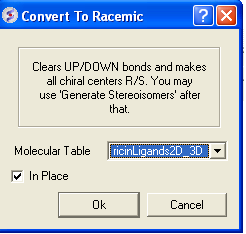
10.20.5 Convert to Racemic |
To remove stereo bonds and make all chemical centers R/S in a chemical table:
- Chemistry/Convert to Racemic
- Select the Molecular Table
- Select In Place if you wish to overwrite the table.
| NOTE: To reassign stereo bonds use the Generate Stereoisomers option |
| Prev List Species at pH | Home Up | Next Build Prediction Model |