| Prev | 2.22 Macros | Next |
[ buildpep | calcBindingEnergy | calcDihedral4atoms | calcDihedralAngle | calcEnsembleAver | calcMaps | calcPepHelicity | calcProtUnfoldingEnergy | calcRmsd | calcSeqContent | icmCavityFinder | dsCellBox | findSymNeighbors | dsCharge | dsChem | dsCustom | dsCustomFull | dsDistance | dsPropertySkin | calcEnergyStrain | icmPmfProfile | dsPrositePdb | dsRebel | dsSeqPdbOutput | dsSkinLabel | dsSkinPocket | dsStackConf | dsVarLabels | ds3D | dsWorm | dsXyz | findFuncMin | findFuncZero | nice | cool | homodel | makeIndexChemDb | makeIndexSwiss | makePdbFromStereo | mkUniqPdbSequences | plot2DSeq | plotSeqDotMatrix | plotSeqDotMatrix2 | plotBestEnergies | plotOldEnergy | plotFlexibility | plotCluster | plotMatrix | plotRama | plotRose | plotSeqProperty | predictSeq | prepSwiss | printFast | printMatrix | printPostScript | printTorsions | refineModel | regul | rdBlastOutput | rdSeqTab | readPdbList | remarkObj | searchPatternDb | searchPatternPdb | searchObjSegment | searchSeqDb | searchSeqPdb | searchSeqFullPdb | searchSeqSwiss | setResLabel | sortSeq | undsCharge | makeSimpleModel | makeSimpleDockObj | searchSeqProsite ]
Macros provide you with a great mechanism to create and develop your ICM environment and adjust it to your own needs (see also How do I customize my ICM environment. ). Very often a repeated series of ICM commands is used for dealing with routine tasks. It is wise not to retype all these commands each time, but rather to combine them into a bunch for submission as a single command. Several examples follow.2.22.1 buildpep: Building peptides from a sequence | [Top] |
creates a new ICM object from an input sequence. This macro recognizes if you specified the sequence in one-letter upper-case letters or lower-case three-letter code and adds uncharged N- and C- termini. Use semicolon ( ; ) to separate molecules. If you want to use different termini, or build a non-peptide molecule apply the build command directly or modify the macro.
For a multimolecular object you can also create separate objects and then move them together.
Examples:
buildpep "ala his trp glu" # one tetrapeptide: nter and cooh added buildpep "ala his ; trp glu" # two di-peptides buildpep "one ; one" # two oxygens buildpep "YTGSNVKVAV" # decapeptide buildpep "AQSVPYGVSQ;IKAPALHSQG" # two decapeptidesThe source code of the buildpep macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.2 calcBindingEnergy: estimates electrostatic, hydrophobic and entropic binding terms | [Top] |
evaluates energy of binding of two complexed molecules ms_1 and ms_2 s_terms for the given set of energy terms s_terms. This macro uses the boundary element algorithm to solve the Poisson equation. The parameters for this macro have been derived in the Schapira, M., Totrov, M., and Abagyan, R. (1999) paper.
Example:
read object s_icmhome+"complex" cool a_ calcBindingEnergy a_1 a_2 "el,sf,en"
The source code of the calcBindingEnergy macro is stored in the $ICMHOME/_rebel file. Feel free to copy and modify the text.
2.22.3 calcDihedral4atoms: calculate a torsion angle defined by four atoms | [Top] |
calculates an angle between the two planes specified by any four atoms, as_1 as_2 as_3 as_4. Usually these are four consequtive covalently bound atoms.
Example:
buildpep "ala his his" display atom label calcDihedral4atoms a_/3/nd1 a_/3/cg a_/3/cd2 a_/3/ne2 Angle= -0.06781 deg. (also saved in) # it is almost flat
The source code of the calcDihedral4atoms macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.4 calcDihedralAngle: calculate an angle between two planes in a molecule | [Top] |
calculates an angle between the two planes specified by two triplets of atoms, specified by the as_plane1 and as_plane2 selections
An example in which we measure an angle between planes of two histidines:
buildpep "ala his his" # we use another macro here display atom labels calcDihedralAngle a_/2/cg,nd1,cd2 a_/3/cg,nd1,cd2 Angle= 131.432612 deg. (in).
The source code of the calcDihedralAngle macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.5 calcEnsembleAver: Boltzmann average the energies of the stack conformations | [Top] |
a macro showing an example of how to calculate a Boltzmann-weighted average given a conformational stack of conformation representatives. The stack may be formed as a result of a Monte Carlo simulation or created manually. The s_parameter string contains any expression returning the parameter to be averaged (e.g. "Value(v_/2/phi)" or "Distance(a_/2/ca a_/4/ca)" ).
Example:
buildpep "ala his his" set vrestraint a_/* # impose rotamer probabilities mncallsMC = 5000 montecarlo # a stack is formed with energies calcEnsembleAver 300. "Value(v_/2/phi)"
See also macro helicity.
The source code of the calcEnsembleAver macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.6 calcMaps: calculate five energy maps and write them to files | [Top] |
calculates five energy grid maps for the current object with the grid size r_gridSize in the 3D box volume defined by the R_box . The maps are saved to files with names s_fileNameRoot_gc.map s_fileNameRoot_gh.map etc. and are deleted upon return from the macro. Be careful with selecting a box. You may focus the box on the area of interest (e.g. Box( a_/55,66 , 7.) ). To use the maps read them in, rename to m_gc m_gh, etc. and set terms "gc,gh,ge,gb,gs" . If you determined the box interactively you may just use the Box () function without arguments (it returns the parameters of the graphical box).
Example:
read object s_icmhome+"crn"
calcMaps "crn" Box( a_/15 4. ) 0.6
read map "crn_ge"
rename m_crn_ge m_ge
display m_ge {1 2 3 0 4 5 6}
# the maps can be used in another session
The source code of the calcMaps macro is stored in the $ICMHOME/_docking file. Feel free to copy and modify the text.
2.22.7 calcPepHelicity: calculate average helicity of a peptide from movie frames | [Top] |
a macro showing an example of how to calculate the helicity of a peptide structure given an ICM movie of the conformations accepted during a Monte Carlo run. A simulation using montecarlo movie option is a prerequisite for this macro. A good script prototype can be found in the $ICMHOME/_folding file. The movie option saves each accepted conformation to a movie file. The secondary structure of all transient conformations is assigned with the assign sstructure command.
Example:
% _folding # run the _folding script with the movie option. % icm read object "mypep" # the name of your peptide object calcPepHelicity "mypep" 600.
See also macro calcEnsembleAver
The source code of the calcPepHelicity macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.8 calcProtUnfoldingEnergy: rough estimate of solvation energy change upon unfolding | [Top] |
calculates an octanol/water transfer solvation energy for the given # conformation as compared to an extended chain conformation.
The source code of the calcProtUnfoldingEnergy macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.9 calcRmsd: calculate three types of Rmsd between protein conformations | [Top] |
calculates Ca-atom, backbone-atom, and heavy-atom RMSD for two input residue selections. The main effort in this macro is to take the internal symmetry of amino-acid sidechains into account.
For example, two phenylalanines related by the 180 degrees rotation of the xi2 angle are identical, but will have a non-zero Rmsd(a_1./phe a_2./phe) because cd1 and ce1 of one selection lay on top of cd2 and ce2 atoms of the second selection, respectively. To calculate this Rmsd correctly, we need to find the rotation The following residues have internal symmetry (or pseudo-symmetry): leu,tyr,phe,asp,glu,arg,val.
The source code of calcRmsd macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.10 calcSeqContent | [Top] |
[ convertObject | clusterChem ]
calcSeqContent s_seqNamePattern ("*")analyzes amino acid composition of the input sequence or sequences. Specify quoted sequence name, pattern (e.g. "*_HUMAN" ) or "*" for all sequences.
Example:
read sequence s_icmhome+"seqs" calcSeqContent "*" # matches names of all sequences .. Statistics for 3 sequence(s): Azur_Alcde Azur_Alcfa Azur_Alcsp AA N % Expected A 42 10.34 7.85 C 9 2.22 2.55 ... calcSeqContent "*de" # sequences ending with 'de' Statistics for 1 sequence(s): Azur_Alcde Res N % Expected A 20 13.42 7.85 C 3 2.01 2.55 D 8 5.37 5.17 E 6 4.03 6.95
The columns are as follows:
- One-letter amino-acid code
- The total occurrence of the amino acid
- Relative percentage occurrence in the given set of sequences
- Expected mean occurrence of the amino acid in proteins
The source code of the calcSeqContent macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
convertObject auto ms_ (a_) l_delete_water (yes) l_optimize_hydrogens (no) l_replace_the_original (no) l_display (no)
converts a non-ICM object into and ICM object and performs some additional refinements. The macro returns r_residialRmsd value containing the Rmsd of the model atoms from the equivalent template atoms (the same value is returned by the convert command in r_out ). If this residual is greater than 0.5 , it usually means some problems with the conversion (e.g. unusual residues, missing parts, etc.).
clusterChem s_inObjects ("*.ob") s_outObject ("clustered.ob")
performs clustering based on chemical similarity
2.22.11 icmCavityFinder: analyze and display cavities | [Top] |
|
calculates and displays cavities in a molecular structure.
These cavities are sorted by size, and displayed.
The l_interactive argument allows cavities to be displayed one by one interactively.
To display the transparent outer shell edit the macro and activate this feature.
The r_minVolume parameters defines the volume of the smallest retained cavity. Increase it if you want only large cavities. For each cavity this macro calculates volume V (in square Angstoms), area A and an effective radius R (compare it with the radius of a water molecule of 1.4A). |
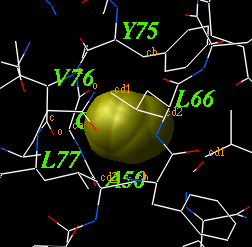
|
The icmCavityFinder macro uses two powerful features of ICM-shell:
- a grob with analytical molecular surface ( a.k.a. skin ) of the selected atoms can be built using the make grob skin as_ as_ "g_skin" command.
- this grob can be divided into the separate grobs with the outer shell and all the inner cavities with the split g_skin command.
- icmCavityFinder also uses the Volume(g) and Area(g) functions to measure volume and area of the cavities, as well as the Sphere( g_ r_radius ) function to select atoms and residues around any grob.
Example:
read object s_icmhome+"1qoc" delete a_w* # remove water molecules icmCavityFinder a_1 yes 4. 3deleted Info> finished surface search, n_of surface atoms = 744 Surface .................................................. Info> finished basic surface element calculations Info> Estimated vertex number = 335800, actual = 184896 Info> packing vertices... sorted... done! Info> skin grob "" created (solid model: 32197 point Info> 3 grobs... Shell 1: V=11039.291805 A=4525.876702 Warning> Volume(created ) may be improperly calculated: env CAVITY 2: V=25.253718 A=44.282805 R~1.710848 -------------- - Num Res. Type ----{SS Molecule}-- Object - sf - sfRati 26 ile Amino I H m 1qoc 0.0 0.00 53 leu Amino L E m 1qoc 0.0 0.00 58 val Amino V _ m 1qoc 0.0 0.00 76 val Amino V E m 1qoc 0.0 0.00 87 val Amino V E m 1qoc 0.0 0.00 89 ile Amino I E m 1qoc 0.0 0.00 ...
The source code of the icmCavityFinder macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.12 dsCellBox: displays crystallographic unit cell | [Top] |
displays unit crystal cell box for the specified object os_ generated according to crystal symmetry parameters. This tiny macro extracts the cell from the object using the Cell function and makes a grob out of this array with the Grobfunction.
macro dsCellBox os_ (a_)
gCell = Grob ("cell" Cell(os_))
display gCell magenta
keep gCell
endmacro
See also: findSymNeighbors
2.22.13 findSymNeighbors: cell and crystallographic neighbors | [Top] |
finds and builds symmetry related molecules around the input selection.
The source code of the findSymNeighbors macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.14 dsCharge: one of many ways to show charge residues | [Top] |
displays CPK representation of positively and negatively charged amino acid residues in red and blue colors, respectively. See also macro undsCharge
macro dsCharge display a_*./asp,glu/o?* cpk red display a_*./lys,arg/nz,n?* cpk blue endmacro
2.22.15 dsChem : chemical style display | [Top] |
3D display of the input atom selection in chemical style and on white background.
If you want to 'flatten' the molecule you can perform a procedure from the following example:
buildpep "trp" # you need an ICM object tzMethod = "z_only" # tether to the z-plane set tether a_ # each atom is tethered to z=0 minimize "tz" # keep the cov. geometry
The source code of the dsChem macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.16 dsCustom: extended display and property-coloring | [Top] |
Displays the specified representation ( "wire", "cpk", "ball", "stick", "xstick", "surface", "ribbon" ) of a molecular selection and colors the selection according to the following series of features:
- atom type ( s_coloringType="atom" ),
- residue type ("residue"),
- unique molecules ("molecule"),
- secondary structure type ("sstructure"),
- N-to-C-terminal chain course (NtoC""),
- B-factors ("bfactor"),
- electric charges ("charge"),
- solvent accessibility ("accessibility"),
- residue polarity ("polarity"),
- residue hydrophobicity ("hydrophobicity")
The source code of the dsCustom macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.17 dsCustomFull macro for molecular display | [Top] |
an extension of the previous dsCustom macro which, in addition, allows to color by an external rarray of 26 elements for each character. This user-defined array may contain any residue property information.
Flag l_areaSelfMode determines if the surface area is calculated for the selection in the context of all the atoms of the object ( no ) or only the selection itself, as if no other atoms existed ( the self mode )
The source code of the dsCustomFull macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.18 dsDistance: display distances between two selections | [Top] |
|
displays distances in a specified range between atoms of
two input atom selections. This macro saves atom names and distances
in T_dist table. This table can later be resorted and analyzed.
Example: read object s_icmhome + "crn" dsDistance a_/15 a_/18 0. 10. show T_dist |
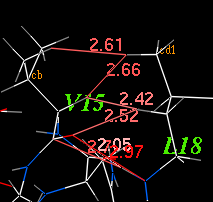
|
The source code of the dsDistance macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.19 dsPropertySkin: display molecular surfaces colored by properties essential for binding | [Top] |
|
dsPropertySkin as_sel (a_) l_wire (yes)
displays essential properties of molecular surfaces which are essential for binding small ligands, peptides or other proteins. The first argument is a selection of atoms involved in the surface calculation. The second argument allows you to display the surface as:
|
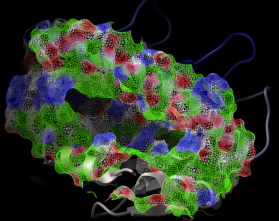
|
The color code:
- white - neutral surface
- green - hydrophobic surface
- red - hydrogen bonding acceptor potential
- blue - hydrogen bonding donor potential
Example shown:
read pdb "1a9e" delete a_w* convert # convert to ICM for map calculations # select receptor atoms 9. away from the peptide with Sphere cool a_ # display ribbon dsPropertySkin Sphere( a_3 a_1 9. ) yes # adjust clipping planes for better effect write image png
Interactive surface display under GUI The same can be performed interactively on ICM objects with the popup-menu:
- display your ICM object
- switch selection level to residue (R)
- select region with selection box or lasso
- click on the right mouse button over one of the selected residues
- selected Display and then Property Skin
- it creates grob g_recSkin which can then be undisplayed and further manipulated
The source code of the dsPropertySkin macro is stored in the $ICMHOME/_docking file. Feel free to copy and modify the text.
2.22.20 calcEnergyStrain: analyzing energy strain in proteins | [Top] |
calculates relative energy of each residue for residue selection rs_ ; and colors the selected residues by strain ( if logical l_colorByStrain is "yes" ). The R_limits argument determines the range represented by the color gradient (i.e. residues strained beyond 5. will still be shown in red).
This macro uses statistics obtained in the Maiorov, Abagyan, 1998 paper.
Example:
read object s_icmhome + "crn" # an ICM object calcEnergyStrain a_/A show ENERGY_STRAIN
The source code of the calcEnergyStrain macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.21 icmPmfProfile | [Top] |
|
macro icmPmfProfile os_ ( a_ ) l_accessibilityCorrection (yes) l_display ( no ) calculates a statistical energy of mean-force for each residue of a provided object. This energy is calculated with the "mf" parameters defined in the icm.pmf file. The residue energies are then normalized to the expected mean and standard deviation of the same residue in real high resolution structures. The mean energy value can be calculated as a function of its solvent accessibility if l_accessibilityCorrection is set to yes. The calculated table contains residue energies and accessibilites. These values can be used to color residues of the molecule according to those values. In an example shown here we build a model (using the build model command of HCV protein on the basis of another viral coat protein. Then the profile was calculated for the model and the original structure. The calculation clearly shows the problematic regions of the model (the red parts) while the source structure looks quite reasonable. |
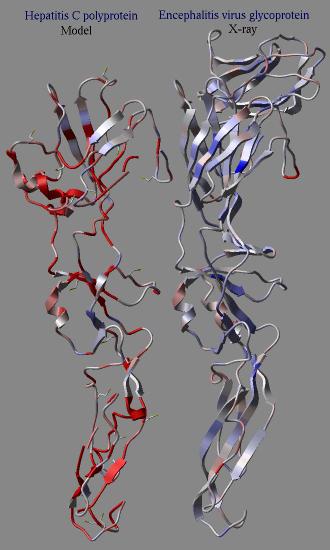
|
2.22.22 dsPrositePdb | [Top] |
Finds all PROSITE pattern-related fragments in the current object and displays/colors the found fragments and residue labels.
The source code of the dsPrositePdb macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.23 dsRebel: surface electrostatic potential | [Top] |
generates the skin representation of the molecular surface colored according to the electrostatic potential calculated by the REBEL method (hydrogen atoms are ignored). The coloring is controlled by the maxColorPotential parameter. This macro uses a simplified charge scheme (a_/lys/nz : 1.0, a_/arg/nh* : 0.5 , a_/asp,glu/oe*,od* : -0.5 ) and uses only the heavy atoms for the calculations for the sake of speed. A full atom version of this macro is dsRebel .
The source code of the dsRebel macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.24 dsSeqPdbOutput : visualize the sequence similarity search results | [Top] |
Goes through a list of PDB hits resulting in find database command and displays alignment(s) of the input sequence(s) with the found PDB structures and SWISSPROT annotations.
The source code of the dsSeqPdbOutput macro is stored in the $ICMHOME/_macro file. Feel free to copy and modify the text.
2.22.25 dsSkinLabel | [Top] |
For all residues specified by the input residue selector, rs_, displays residue labels shifted toward the user to make the labels visible when skin representation is used.
2.22.26 dsSkinPocket and dsSkinPocketIcm | [Top] |
display the receptor pocket around the selected ligand ms_ligand. Only the largest contiguous pocket surrounding the ligand is retained for clarity. The dsSkinPocketIcm macro also colors the molecular surface by hydrogen bonding potential and hydrophobicity. Best used with the ligand shown in cpk, if the ligand is small.
These macros can also be used to show the protein-protein interface.
Example:
read object s_icmhome+"complex" cool a_ dsSkinPocket a_1 a_2 7. # shows the surface of a_1
2.22.27 dsStackConf | [Top] |
displays superimposed set of conformations from a conformational stack for given selection as_.
2.22.28 dsVarLabels | [Top] |
displays color labels for different types of torsion variables.
2.22.29 ds3D | [Top] |
|
ds3D M_interObjectDistances [S_names]
display 3D coordinates corresponding to an input square distance data matrix. Relative errors (in percent) of embedding to 3D space are in R_out: first entry is for the total error, next three are for X, Y and Z coordinates. |
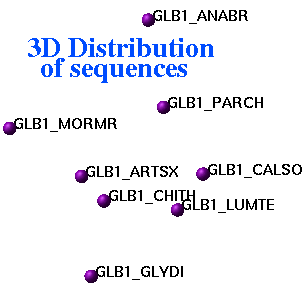
|
| Representation of inter-sequence evolutionary distances in three-dimensional space |
2.22.30 dsWorm | [Top] |
displays "worm" (or "tube") representation of selected molecule(s). Residue colors are smoothly changed from blue (at N-terminus) to red (at C-terminus).
2.22.31 dsXyz : display | [Top] |
 displays points from the N_atoms x 3 matrix of M_xyz_coordinates
in 3D space as blue balls.
The origin of the Nx3 matrix is not important.
The macro creates an object called a_dots.
In this object each dot is a one-atom residue called 'dot'. The atom type
is arbitrarily assigned to oxygen, and the atom names are 'o'.
displays points from the N_atoms x 3 matrix of M_xyz_coordinates
in 3D space as blue balls.
The origin of the Nx3 matrix is not important.
The macro creates an object called a_dots.
In this object each dot is a one-atom residue called 'dot'. The atom type
is arbitrarily assigned to oxygen, and the atom names are 'o'.
One can further manipulate this object, e.g. color a_/12:15/o green .
An example in which we generate sparse surface points at vwExpand distance around a molecule and display them.
buildpep "ala his trp" mxyz = Xyz( a_ 5. surface ) display skin white dsXyz mxyz color a_dots. red
2.22.32 findFuncMin | [Top] |
minimizes one-dimensional functions provided as a string with the function expression. The macro uses successive subdivision method, and assumes that the function derivative is smooth and has only one solution in the interval
Example:
findFuncMin "Sin(x)*x-1." , -1. 2. 0.00001 -1.000000 < x < 0.500000 -0.250000 < x < 0.500000 -0.062500 < x < 0.125000 .... -0.000004 < x < 0.000008 -0.000004 < x < 0.000002
2.22.33 findFuncZero | [Top] |
finds a root of the provided function of one variable with specified brackets with iterations. E.g.
findFuncZero "x*x*x-3.*x*x" 1. 33. 0.00001 => x=17.000000 F=4046.000000 => x=9.000000 F=486.000000 => x=5.000000 F=50.000000 => x=3.000000 F=0.000000
2.22.34 nice | [Top] |
reads and displays a PDB structure in ribbon representation; colors each molecule of the structure by colors smoothly changing from blue (at N-terminus) to red (at C-terminus).
Example:
nice "365d" # new DNA drug prototype nice "334d" # lexitropsin, derivative of netropsin
2.22.35 cool | [Top] |
similar to the macro nice above, but refers to a residue selection.
2.22.36 homodel | [Top] |
homology modeling macro. The first sequence in the input alignment should contain the sequence of a PDB template to which the modeling will be performed. If flag l_quick is on, only an approximate geometrical model building is performed. You can also use the build model command directly.
2.22.37 makeIndexChemDb | [Top] |
Creates and saves an index to a small compound database existing in standard mol or mol2 formats (specified by the s_dbTypeparameter). s_dbIndex defines full-path root name of several index-related files. String array S_dbFields specifies fields of the input database which are indexed by the macro.
An example in which we index the cdi.sdf file and generate the cdi.inx file in a different directory:
% icm
makeIndexChemDb "/data/chem/chemdiv/cdi.sdf" "/data/icm/inx/cdi" "mol" {"ID"}
2.22.38 makeIndexSwiss | [Top] |
Creates and saves an index to the SWISSPROT sequence database (datafile s_swiss). s_indexName defines the root name of several index-related files with respect to ICM user directory, s_userDir.
2.22.39 makePdbFromStereo: restore 3D coordinates from a stereo picture | [Top] |
2.22.40 mkUniqPdbSequences | [Top] |
Creates a collection of PDB sequences with specified degree of mutual dissimilarity, i_percentDifference. Replace old dataset if l_replace is on.
2.22.41 plot2DSeq | [Top] |
generates a 2D representation of "distances" between each pair of sequences from the input alignment.
2.22.42 plotSeqDotMatrix | [Top] |
generates an EPS file in which local sequence similarities between two sequences are shown in the form of a two-dimensional dot-matrix plot. Significance of local sequence similarities is shown by logarithm of the probability values and is calculated in multiple windows from i_mi to i_mx. The log-probability values are color-coded as follows: light blue: 0.7, red 1.0.
2.22.43 plotSeqDotMatrix2 | [Top] |
generates an EPS file in which local sequence similarities between two sequences are shown in the form of a two-dimensional dot-matrix plot. Significance of local sequence similarities is shown by ( 1. - Probability(..) ) values and is calculated in multiple windows from i_mi to i_mx. The ( 1 - P ) values are color-coded as follows: light blue: 0.7, red 0.99.
2.22.44 plotBestEnergy | [Top] |
plots profile of energy improvement during an ICM Monte Carlo simulation. Data are taken from the MC output log file or files, s_McOutputFile. You can specify a single output file (e.g. "f1.results"), or several files, e.g. "f1.ou,f2.ou", or drop the default ".ou" extension, e.g. "f1,f2,f2".
This macro gives you an idea about the convergence between several runs.
2.22.45 plotOldEnergy | [Top] |
plots profile of energy changes during an ICM Monte Carlo simulation. Data are taken from the MC output log file, s_McOutputFile.
2.22.46 plotFlexibility | [Top] |
calculates and plots flexibility profile for input sequence seq_ and smooths the profile with i_windowSize residue window.
2.22.47 plotCluster | [Top] |
plot distribution of clusters. Arguments:
- a square matrix of distances between n objects. For arrays it may be calculated with the
Distance ( ) function (e.g. Distance(Xyz(a_//ca))). For angular RMSD the
distance can be calculated from a matrix v of values of torsion angles for many conformations:
for i=1,n-1 di[i,i]=0. for j=i+1,n # takes care of -179 and 181, base 360 is the default dif=Remainder(v[i]-v[j]) # angular RMSD di[i,j]=Sqrt(Mean(dif*dif)) di[j,i]=di[i,j] endfor endfor di[n,n]=0. -
sarray of names for each of n points . Possibilities:
- the empty sarray: {""}. No name tags will be attached to each point
- Sarray(Count(n)) generates names like this: {"1" "2" "3" ... }
- user-defined: e.g. Name(a_*.) if each point correspond to an object
- manual: e.g. {"A" "compound X" "c"}
- See also: arguments for the plotcommand.
2.22.48 plotMatrix | [Top] |
generates combined X-Y plot of several Ys (2nd, 3rd , etc. rows of the input matrix M_data) versus the one X-coordinate, assumed to be the first row of the matrix. i_numberPerLine parameter defines the size of the plotted block size if the number of data points is greater then i_numberPerLine. i_orientation equal to 1 defines portrait orientation of the output plot, landscape otherwise.
2.22.49 plotRama | [Top] |
generates a phi-psi Ramachandran plot of an rs_ residue selection. If logical l_show_residue_label is on, the macro marks the residue labels. If l_shaded_boundaries is on, the allowed (more exactly, core) regions are shown as shaded areas; otherwise the contours of the core regions are drawn.
2.22.50 plotRose | [Top] |
just a nice example of a simple macro generating "rose" plot.
2.22.51 plotSeqProperty | [Top] |
a generic macro to plot local sequence properties. Modify it for your convenience. Here is an example in which we plot residue b-factors along with the crambin sequence. s_seqString could be the sequence (e.g. String(1crn_m) ) or secondary structure, (e.g. Sstructure(1crn_m) ) or any other string of the same length as the sequence.
read pdb "1crn"
make sequence
b = Bfactor( a_/* )
plotSeqProperty b String(1crn_m ) {"" "" ""} "tm.eps" 20 "portrait"
2.22.52 predictSeq | [Top] |
calculates and plots hydrophobicity and flexibility profiles and secondary structure diagrams for the given sequence s_seq (this is a string with the sequence name) and saves the results in s_fileName PostScript file.
2.22.53 prepSwiss | [Top] |
extracts all sequences from the SWISSPROT database which exclude ( l_exclude= yes ) or include ( l_exclude= no) the specified sequence pattern, s_IDpattern and creates a set of database files with the rootname s_file intended to use in the command find database.
2.22.54 printFast | [Top] |
writes current content of the graphic window to a PostScript file and calls unix ' lp' command to send the resulting PostScript file to the specified printer, s_ofPrinterName .
2.22.55 printMatrix | [Top] |
prints matrix M_matrix according to the input format s_format.
2.22.56 printPostScript | [Top] |
converts the current content of the graphics window to a PostScript file and directs it to the s_ofPrinterName printer.
2.22.57 printTorsions | [Top] |
outputs all torsion angles of the input residue selection.
2.22.58 refineModel: globally optimize side-chains and anneal the backbone | [Top] |
This macro can be used to improve any ICM model. The model can come from the build model command or the convert command or regul macro, etc. It performs
- side-chain sampling using montecarlo fast
- interative annealing with tethers you have provided
- second side-chain sampling to resolve the new problems resulting from the second step
2.22.59 regul | [Top] |
creates a regularized ICM-model of an input residue selection ( rs_ ) under the name s_regObjName. If l_newIcmSeq is set to yes , the macro will create sequence from that of the input residue selection, optionally modified by the N- and C-terminal groups ( s_ngroup and s_cgroup, empty "" strings are allowed); otherwise the macro will use an ICM-sequence file, s_regObjName.se The protocol course may be displayed if l_displayRegul set to yes . The resulting ICM model will be written to file s_regObjName.ob. If l_freeMin set to *yes*, the resulting model will be additionally minimized, now without tethers, and be written to file s_regObjNamef.ob. The summary of the macro's work will be saved to file s_regObjName.log .
2.22.60 rdBlastOutput | [Top] |
reads a set of sequences defined in a BLAST's output file, S_giArray from the NCBI database.
2.22.61 rdSeqTab | [Top] |
reads a set of sequences listed in the ICM-table SR, an output of find database command, from the database defined by s_dbase.
2.22.62 readPdbList | [Top] |
reads a series of PDB files specified in the input string array and creates sequences for all loaded structures.
2.22.63 remarkObj | [Top] |
allows editing an annotation (comment) of the current object. Existing comment (if any) is read in an editor and after modification assigned to the object.
2.22.64 searchPatternDb | [Top] |
searches for the pattern in the sequences of the specified indexed database s_dbase.
2.22.65 searchPatternPdb | [Top] |
searches for the specified pattern in pdb sequences taken from the foldbank.db file.
Example (first hydrophobic residue, then from 115 to 128 of any residues, non-proline and alanine at the C-terminus):
searchPatternPdb "^[LIVAFM]?\{115,128\}[!P]A$"
2.22.66 searchObjSegment | [Top] |
for given molecule ms_ finds all examples of similar 3D motifs not shorter than i_MinNofMatchingResidues residues with the accuracy r_RMSD A in the ICM protein fold database.
2.22.67 searchSeqDb | [Top] |
search the database s_dbase using query sequence(s) specified in S_seqNames. Found hits and their specs are collected in the output table file s_projName.tab. If logical flag l_appendProj is on data will be appended to the existing table. Similarity of hits to the query sequence(s) is controlled by parameter r_probability (see Probability()).
2.22.68 searchSeqPdb | [Top] |
sequence search of all currently loaded sequences in the sequences of the proteins from the fold bank collection. Found hits and their specs are collected in the output table file s_projName. If logical flag l_appendProj is on data will be appended to the existing table. Similarity of hits to the query sequence(s) is controlled by parameter r_probability (see Probability()).
2.22.69 searchSeqPdb | [Top] |
sequence search of all currently loaded sequences through all proteins from the collection s_pdbDir+"/derived_data/pdb_seqres.txt.Z", a subset of PDB sequences with given degree of mutual dissimilarity. Found hits and their specs are collected in the output table file s_projName. If logical flag l_appendProj is on data will be appended to the existing table. Similarity of hits to the query sequence(s) is controlled by parameter r_probability (see Probability()).
2.22.70 searchSeqSwiss | [Top] |
Searches for homologues of the query sequence seq_ in the SWISSPROT database.
2.22.71 setResLabel | [Top] |
moves displayed atom labels to the atoms specific to each residue type.
2.22.72 sortSeq | [Top] |
sort sequences by their length and suggest outliers.
2.22.73 undsCharge | [Top] |
color display of the charged residues.
2.22.74 makeSimpleModel | [Top] |
This macro rapidly builds a model by homology using simplified residues described in the residue library. Input data are the sequence of the model, seq_ and alignment ali_ of the model's sequence with the template object os_ .
2.22.75 makeSimpleDockObj | [Top] |
This macro builds an ICM object from simplified residues described in the residue library. The goal is to convert an all-atom molecular object into an object in simplified representation for fast docking calculations.
2.22.76 searchSeqProsite | [Top] |
compares input sequence against all sequence patterns collected in the PROSITE database.
Examples:
read sequence "zincFing.seq" # load sequences
find prosite 2drp_d # search all < 1000 patterns
# through the sequence
find profile 2drp_d # search profile from prosite database
See also:
find pattern, find database pattern=s_pattrn, find prosite.
| Prev Xyz | Home Up | Next files |