| Prev | 2.19 Other shell variables | Next |
[ defCell | accFunction | gapFunction | _I_out | M_out | _R_out | _S_out | swissFields | readMolNames | sv | as_out | as2_out | vv | vs_out ]
2.19.1 defCell | [Top] |
Default: {1. 1. 1. 90. 90. 90.}
2.19.2 accFunction | [Top] |
|
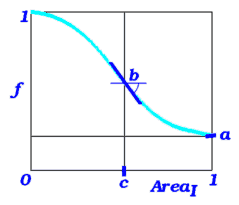
the real array of the solvent accessibility penalty parameters (as described in Batalov and Abagyan, 1999). It contains the values of a, b, c and E damping parameters for aminoacid substitution scores. Generally, if a residue is completely buried ( Area=0), its substitution scores will be used without changes. If it is completely exposed, its substitution scores will be multiplied by the minimal possible value of a. Between these cases the substitution scores are modulated by a smooth ("arctangent") function with a saddle point at Area=c, where the slope will be -b. The fourth parameter is reserved for development. |
This definition is effectively implemented in the Align seq_1 seq_2 area ) }, Score functions and find database command.
Default: {0.33 2.35 0.211, -15.0}.
See also: alignMethod .
2.19.3 gapFunction | [Top] |
ATTENTION: at the present time this gapFunction is only active when alignMethod =2.
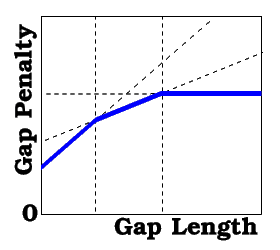
The first two values replace gapOpen and gapExtension traditional values. If present, the third element of the array represents the length of the gap, starting at which further gapExtensions become equal to the fourth element of the array. Likewise, if more elements are present, they represent pairs of the threshold lengths of the gap and the new gapExtensions values. For example,
gapFunction = {2.4 0.15 10. 0.05 20. 0.}
means that
- gap penalty=2.25+0.15*L for L={ 0..10} (and for L=1 it is 2.4= gapOpen ),
- gap penalty=3.25+0.05*L for L={11..20} and
- gap penalty=4.25 for L>20
|
The calculations are fastest for the traditional two-element gapFunction. The three- or four-element gapFunction invokes the optimized routines and is 50-70% slower. The general kind gapFunction costs approximately 70-90% additional time for every pair of gapFunction values. If the last gapExtension is zero, it may be omitted. This definition is effectively implemented in the Align, Score functions and find database search command. |
Default: {2.4 0.15}.
Recommended (put it in your _startup file): gapFunction = {2.4 0.15 10.}
This set will produce fast and structure-like alignments.
See also alignMethod, and accFunction (the accessibility attenuation parameters).
2.19.4 I_out | [Top] |
2.19.5 M_out | [Top] |
2.19.6 R_out | [Top] |
2.19.7 S_out | [Top] |
2.19.8 swissFields | [Top] |
2.19.9 readMolNames | [Top] |
... M END > <CAT_NO> R150002 > <NAME> (5-OXO-HEXAHYDRO-PYRANO[3,2-B]PYRROL-1-YL)-ACETIC ACID METHYL ESTER $$$$In this particular case before using such database set
readMolNames = {"<CAT_NO>" "<NAME>"} # useful for Sigma-Aldrich files
Another example:
readMolNames = {"<CODE>" "<IUPAC_NAME>"} # useful for ACD database
2.19.10 Named Atom/Residue/Molecule/Object Selections | [Top] |
cc = a_//ca # created named selection variable cc show cc & a_/3:15 # use it in the expressionIn this case the named selection cc is a true ICM-shell variable, not just an alias for the Ca selection. Please do not confuse it with another useful mechanism which allows you to use a string in a selection. This mechanism is used in scripts and macros.
Example:
cc = "a_//ca" # in this case cc is a string, not a selection show $cc & a_/3:15 # $cc is replaced by a_//ca before parsing
2.19.11 as_out | [Top] |
2.19.12 as2_out | [Top] |
2.19.13 Named Selections of Internal Variables (Dihedrals, Angles and Bonds) | [Top] |
2.19.14 vs_out | [Top] |
- read variable saves a selection of loaded variables;
| Prev WEBAUTOLINK | Home Up | Next icmcommands |