| Prev | ICM User's Guide 16.22 R-Group Decomposition | Next |
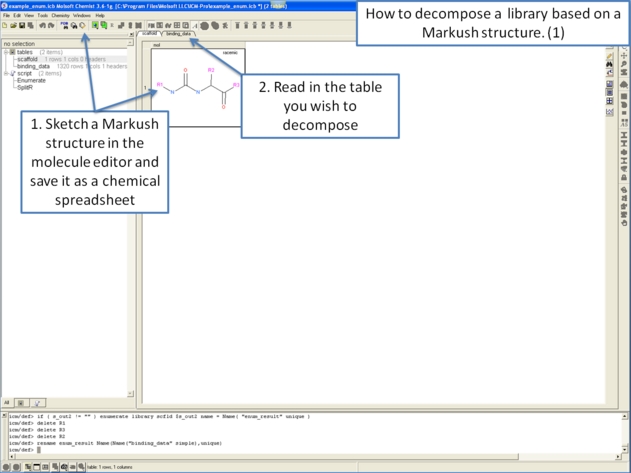
To decompose a library into fragments based on a Markush scaffold (opposite of R-group (Markush) enumeration):
- Read the sdf file you wish to decompose into ICM and it will be displayed as a molecular table.
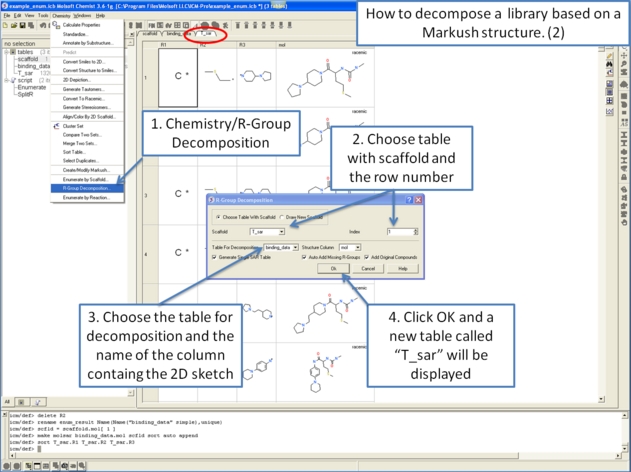
- Chemistry/R-Group Decomposition and a window as shown below will be displayed.
- You now have two options on how to define the Markush scaffold. You can either 1). Draw it using the molecular editor and the smiles string will be added to the window shown below or 2). select a row of a prexisting table.
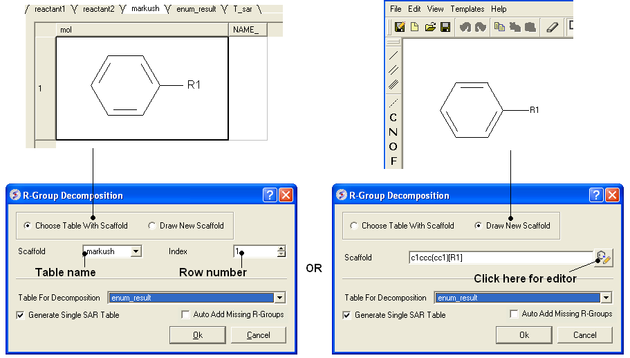
- Use the drop-down option to select the table you wish to decompose.
- If you have more than one R-group ICM can either generate a different table for each R-group or it can merge it into one single table whereby column will represent R1 and column two R2 .... This option is useful if you want to generate a SAR table with a column of activity data next to the R1 and R2 columns (see below).
- If you check the box "Auto Add Missing R Groups" then unique R-groups will be extracted from the scaffold where hydrogens can be attached.
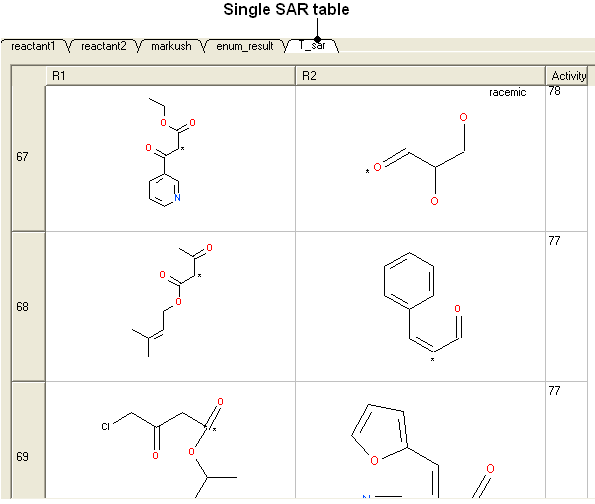
16.22.1 How to decompose a library based on a Markush structure. |
| Prev Enumerate Markush | Home Up | Next Enumerate by Reaction |