| Prev | ICM User's Guide 16.11 Generate Tautomers | Next |
Theory
Tautomers are formed by an interconvertible reaction called tautomerization whereby there is a formal migration of a hydrogen atom along with a switch of a single bond and an adjacent double bond. A common example is the keto to enol tautomerism:
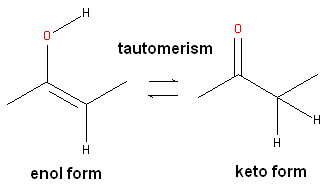
During tautomerization a chemical equilibrium of the tautomers will be reached based on several factors, including, pH, temperature and solvent. Tautomerizations are catalyzed by: bases (deprotonation, formation of a delocalized anion, and, protonation at a different position of the anion; and acids (protonation, formation of a delocalized cation, and deprotonation at a different position adjacent to the cation).
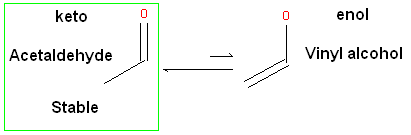
ICM will only generate energetically favorable tautomers. Generally tautomers that have a change in hybridization state are less stable and so ICM will not generate these thus reducing the number of scaffolds generated. For example the keto form shown below is more stable by ~14 kcal.mol than the enol therefore ICM will not generate the enol form.
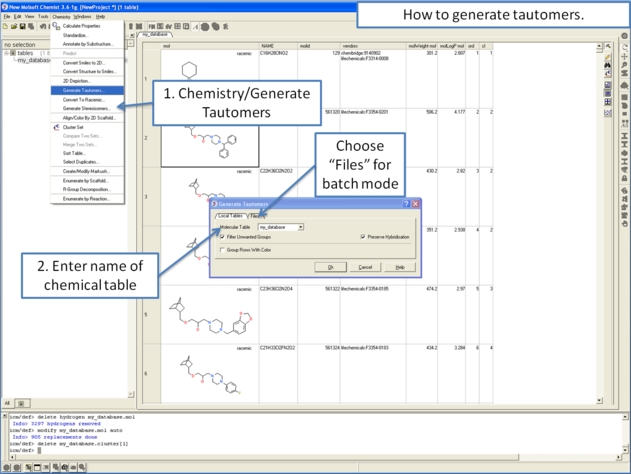
To generate tautomeric conformations of your compound:
- Select the compound(s) in the molecular table. Selected compounds are highlighted in blue.
- Right click in the table and select the option Chemistry/ Enumerate Tautomers. Or select the Chemistry menu/Generate Tautomers.
- If you select Chemistry/Generate Tautomers a dialog box will be displayed. There is also an option to run in batch mode (click the Files Tab).
- Choose the table containing the compounds using the drop-down list.
- Filter Unwanted Groups option will filter results from patterns in the TAUTOFILTER.tab file provided in the distribution. If results match any row from that table then the it will be excluded.
- Preserve Hybridisation Although generally a change in hybridisation state will generate less stable compounds in some cases this is not the case and so you can choose to change hybridisation for a single atom.
- Group Rows With Color option will color tautomers from the same compound with the same color to visually highlight groups.
- The compounds will be displayed in a separate molecular table called TAUTOMERS.
| Prev Generate 3D Conformers | Home Up | Next Convert to Racemic |