| Prev | ICM User's Guide 16.9 Convert to 3D | Next |
[ Convert PDB Chemical | ICM Workspace | Graphical Display ]
To convert a chemical structure from 2D to 3D:
There are three ways in which to do this depending on whether you have a chemical in a chemical table or in the ICM workspace.
From the Chemistry/Convert To 3D option from a table.
- Select the table from the drop down list.
- Select to keep hydrogens and/or fix amide bonds.
- Keep current table or overwrite.
If the compound is in the ICM Workspace:
- Select all of the structure to be converted by double clicking on it in the ICM workspace or by using other selection tools described in the Selection Toolbar Section of this manual.
- MolMechanics/ICM-Convert/Chemical and a data entry box as shown below will be displayed.
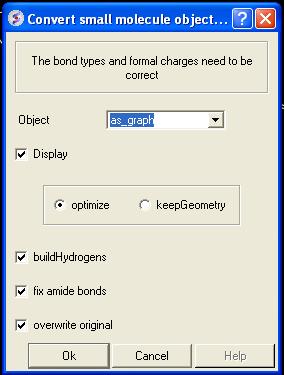
If you have selected the compound as described above the "as_graph" option in the Object data entry box will suffice. You can decide whether you wish to keep the chemical geometry or optimize it in a force-field. Other options include whether you wish to add hydrogens and fix amide bonds.
From an ICM chemical table:
- Select which structures you wish to convert in the molecular table. For instructions on making selections within tables see the Making Table Selections part of this manual.
- Right click on one of the selections you have made and a menu as shown below will be displayed. Or use the Chemistry menu and select Convert To 3D.
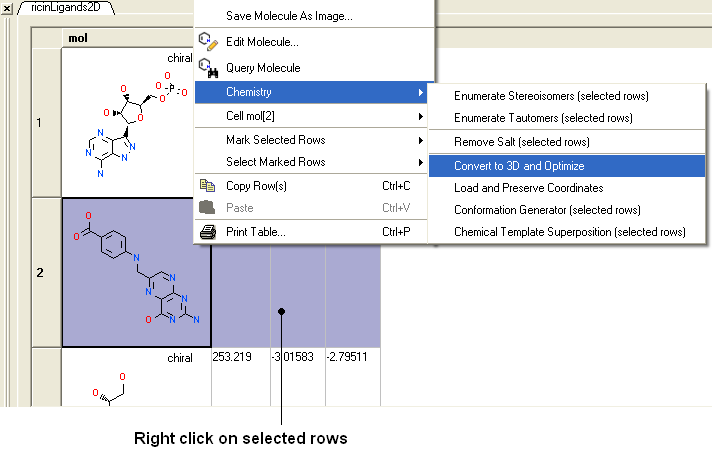
- Select the Chemistry/Convert to 3D and Optimize option and you will see the compounds being converted and minimized in the 3D graphical display window.
Once converted the compounds will be displayed in the 3D graphical display window and also in the ICM workspace.
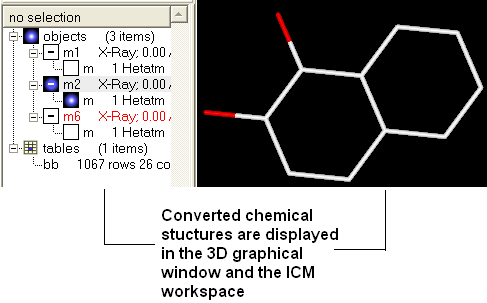
Another way to convert all the ligands contained within a table (or a selection) into 3D coordinates :
- Chemistry/Convert to 3D..
- Use the drop down list to select the Molecular Table.
- Select whether you want to Keep Hydrogens, Fix Omegas and In Place. Select In Place if you want to overwrite current table.
- Click OK
- If you wish to run in Batch mode select the Files option.
| NOTE: Use the 3D-Browse mode to view the chemicals in the graphical display. |
To convert 3D representation in a molecular table back to 2D:
- Chemistry/2D depiction
- Use the drop down list to select the Molecular Table.
- Select In Place if you want to overwrite the current table.
- Click OK.
16.9.1 Converting a Chemical from the PDB |
The protein data bank has not been storing any information about covalent bond types and formal charges of the chemical compounds interacting with proteins! This oversight makes it impossible to automatically convert those molecules to anything sensible and requires your manual interactive assignment of bond types and formal charges for each compound in a pdb-entry. Therefore, if you apply the convert command to a pdb-entry with ligands, the ligands will just become some crippled incomplete molecules that can not be further conformationally optimized.
Therefore, follow these steps to convert a chemical properly from a pdb form to a correct icm object. There are two ways to do this either via the ICM Workspace (recommended) or via the Graphical Display.
16.9.2 Converting a Chemical from the PDB using the ICM Workspace |
- File/Open PDB
- View the ligand in the ICM Workspace by expanding the molecule tree (see below).
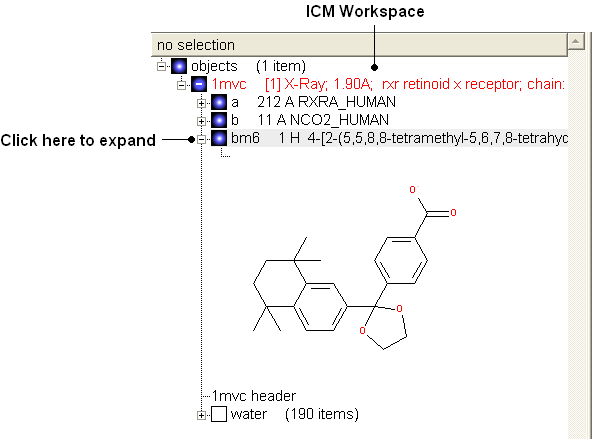
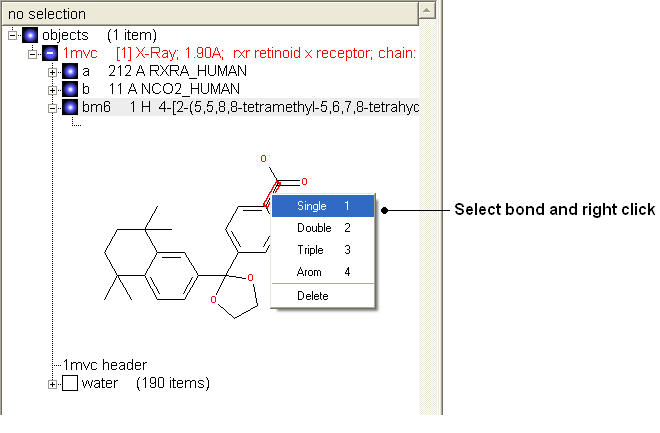
Change bond orders:
- Change the bond orders by selecting the bond (highlighted in red).
- Right click and select the desired bond as shown below.
| NOTE: Keyboard shortcuts are provided to make editing faster. |
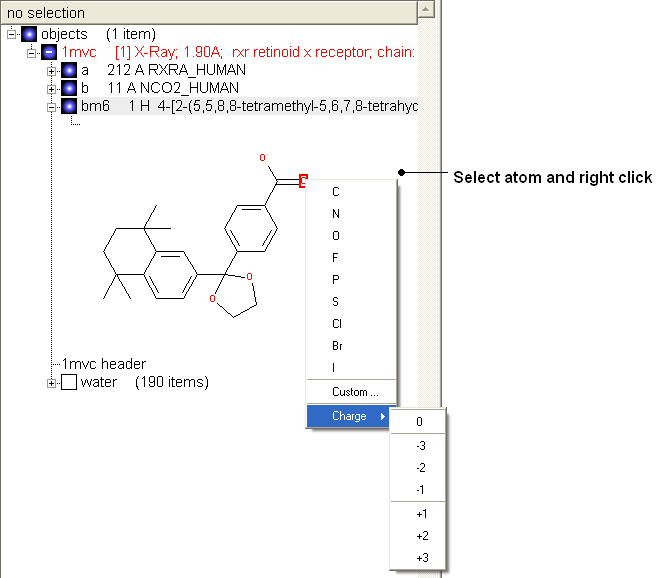
Change atom and charge:
- Change the atom or charge by selecting the atom (highlighted in red).
- Right ckick and select the desired atom or charge as shown below.
| NOTE: Keyboard shortcuts are provided to make editing faster. |
Convert to 3D in MMFF force field:
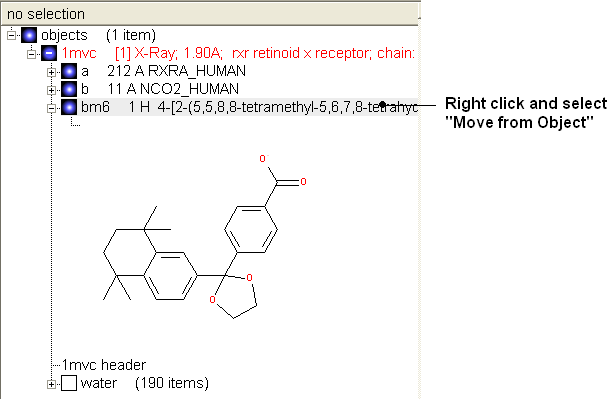
- Once you have made the changes to the ligand - right click on the name of the ligand in the ICM Workspace and select Move from Object.
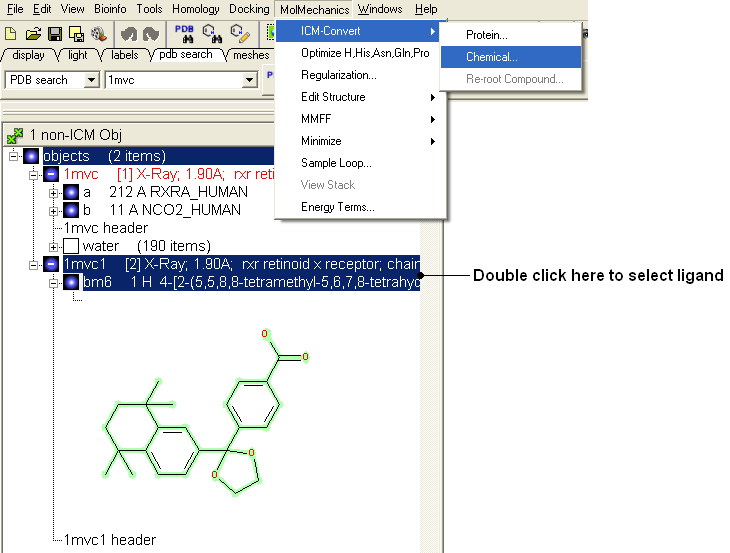
- Select the ligand by double clicking on it in the ICM Workspace.
- Select MolMechanics/ICM-Convert/Chemical
| NOTE: If you need to add an extra bond you will need to use the full molecular editor. Right click on the name of the ligand in the ICM Workspace and select Edit/Edit Compound. |
16.9.3 Converting a Chemical from the PDB using the Graphical Display |
- Display the molecule in wire chemistry style mode by right clicking on the Wire Representation button (see Wire Representation section).
To change the bond types in your ligand:
- Click on MolMechanics/Edit Structure/Set Bond Type and the Set chemical bond type data entry box will be displayed.
You can either select (see selection menu section)the atoms you wish to change graphically using the rectangular or lasoo selection button OR
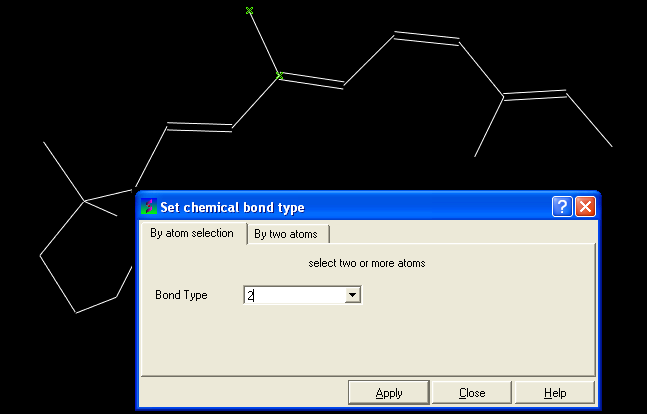
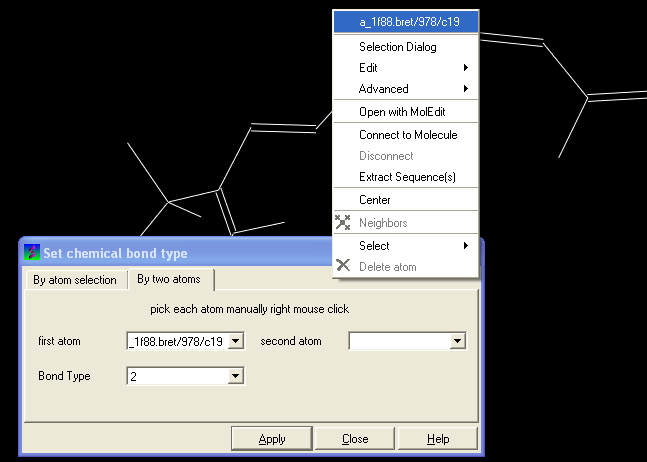
You can select the By two atoms tabs and right click on the atoms you wish to change and then selecting the atom descriptor with the left mouse button as shown below.
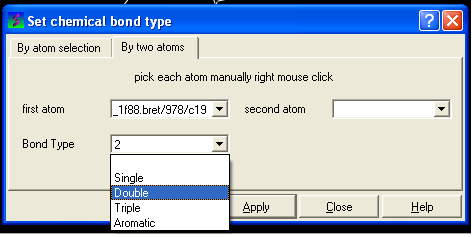
- Select the desired bond type either single, double, triple or aromatic.
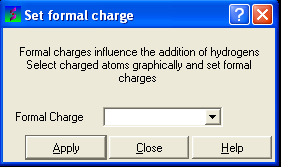
To set the formal charge of a compound:
Click on MolMechanics/Edit Structure/Set Formal Charge and then select the appropriate charge.
The final step is to convert the compound into an ICM object:
- Select the chemical (green crosses in graphical display).
- MolMechanics/ICM-Convert/Chemical
| Prev 2D Depiction | Home Up | Next Generate 3D Conformers |