| Prev | ICM User's Guide 16.19 Select Duplicates | Next |
| NOTE: Gui option is available in versions 3.5-1o and higher. The command line options for this function are described in the ICM Command Language manual at http://www.molsoft.com/man/icm-functions.html#Index-chemical |
This option allows you to select and remove duplicate chemicals in a table.
- Read a chemical table into ICM.
- Chemistry/Select Duplicates
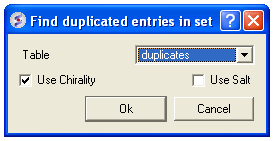
- Enter the table name you want to check for duplicates
- Enter whether you want chirality or the salts included in the analysis.
- Press OK
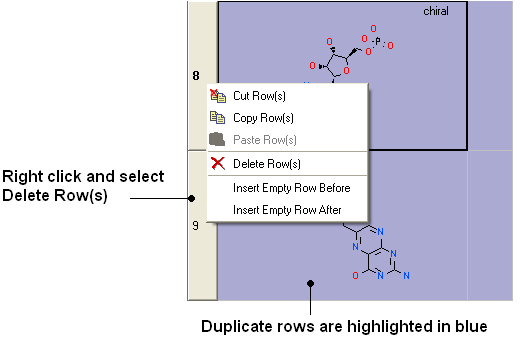
- Duplicate compounds will be highlighted blue in the table. You can delete them by right clicking on the row header ans selecing Delete Rows(s)
| Prev Sort Table | Home Up | Next Markush |