| Prev | ICM User's Guide 22.12 How to Work with the ICM 3D Ligand Editor | Next |
[ Setup Ligand | Setup Receptor | Preferences | Default Display | Surface | Hydrogen Bonds | Energy Circles | Display Hydrogens | Unsatisfied Hydrogen Bonds | Center Ligand | Edit | Undo/Redo | Add Substituent | Multiple Substitutents | Edit 2D | SCORE and Strain | Save Spreadsheet | Pocket Size | Minimization | Dock | Restrain | Screen Substituents | Fragment Linkers ]
22.12.1 How to setup the ligand in the ICM 3D Ligand Editor. |
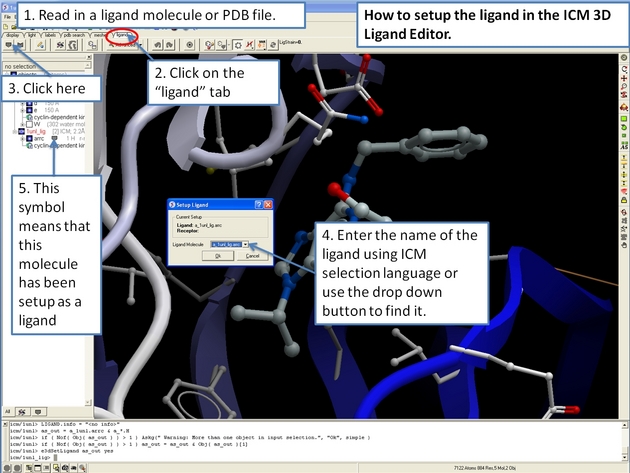
22.12.2 How to setup the receptor in the ICM 3D Ligand Editor. |
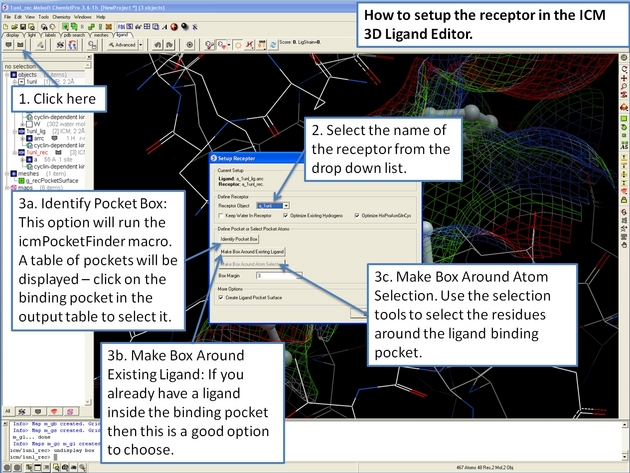
22.12.3 How to change the 3D Ligand Editor preferences. |
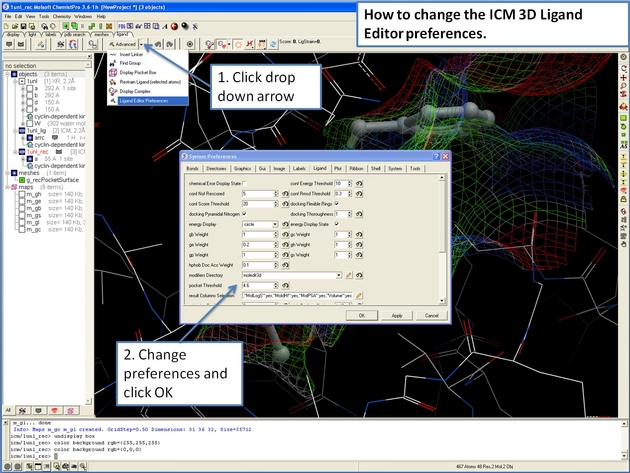
22.12.4 How to configure the default display in the ICM 3D Ligand Editor. |
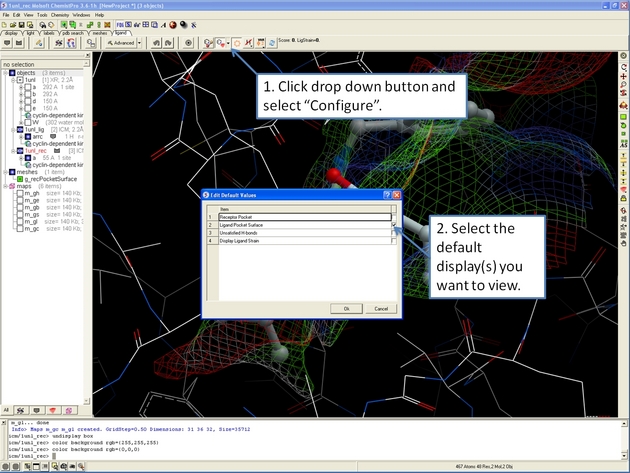
22.12.5 How to display and undisplay the ligand surface representation in the ICM 3D Ligand Editor. |
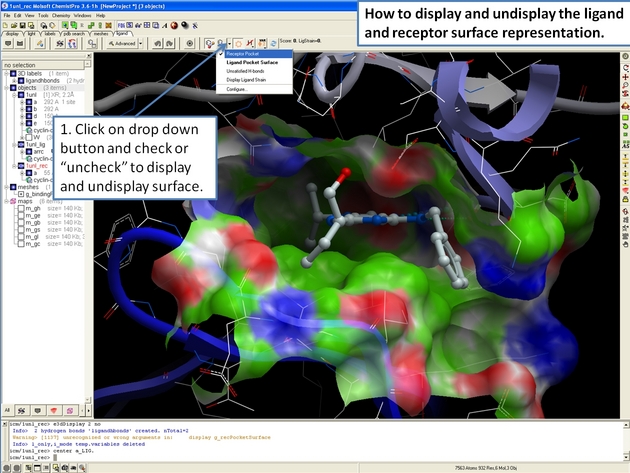
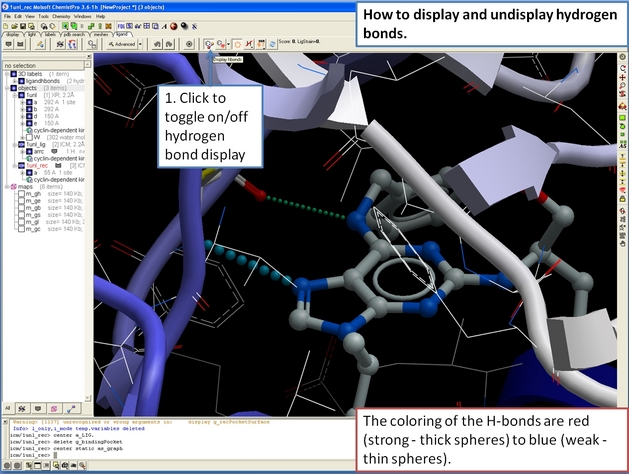
22.12.6 How to display hydrogen bonds in the ICM 3D ligand editor. |
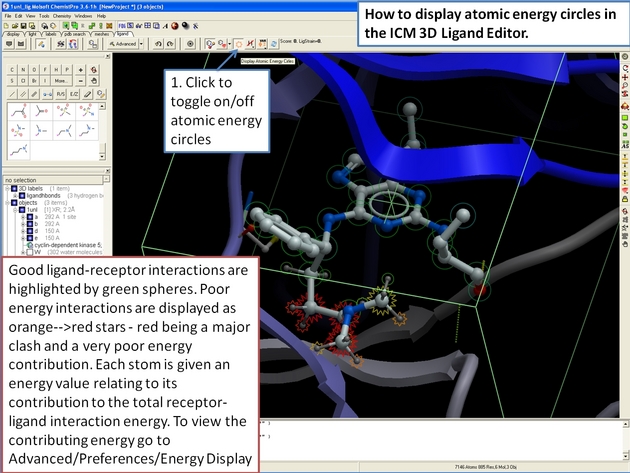
22.12.7 How to display energy atomic circles in the ICM 3D Ligand Editor. |
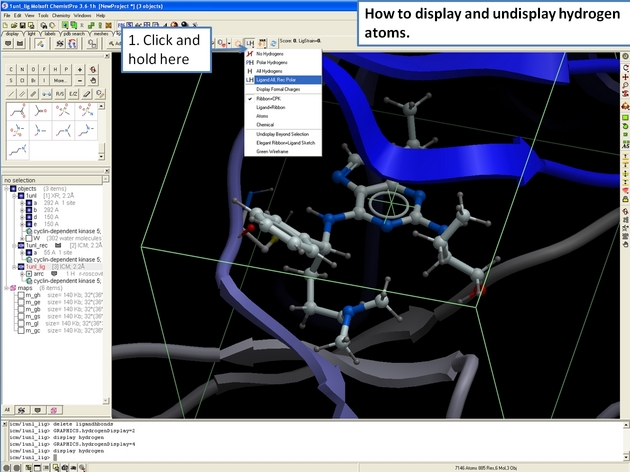
22.12.8 How to display and undisplay hydrogen atoms in the ICM 3D Ligand Editor. |
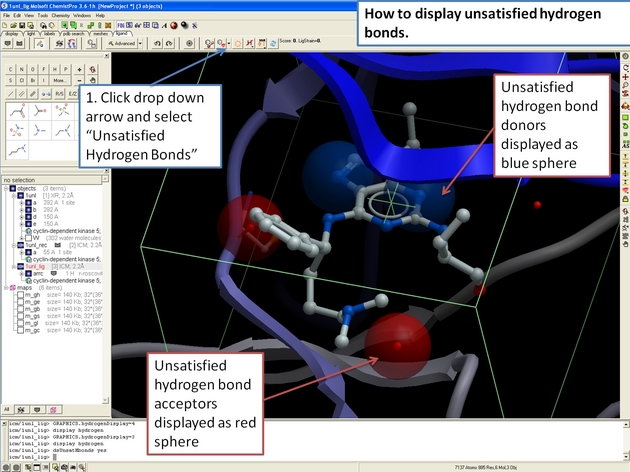
22.12.9 How to display unsatisfied hydrogen bonds in the ICM 3D Ligand Editor. |
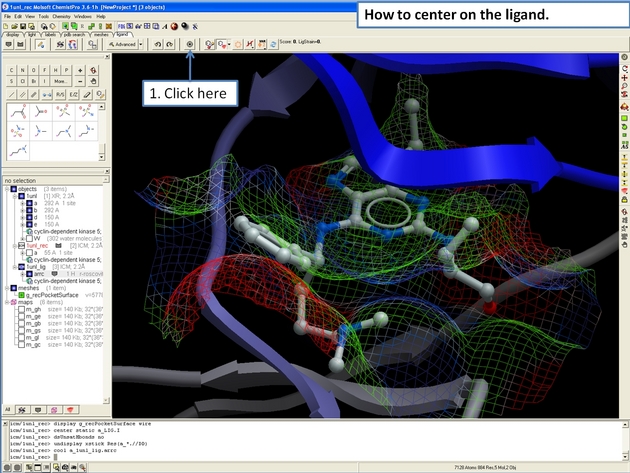
22.12.10 How to center on a ligand in the ICM 3D Ligand Editor. |
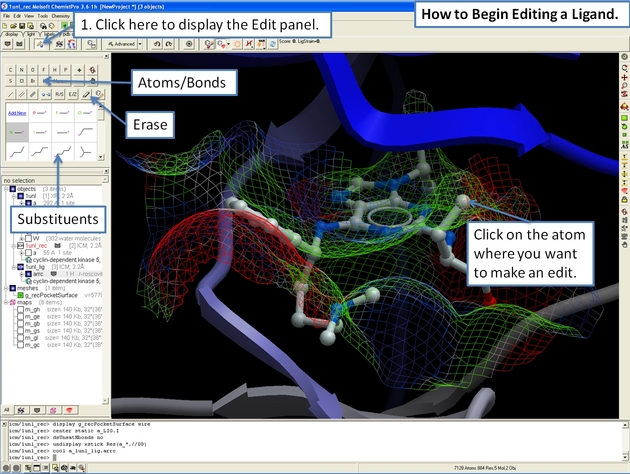
22.12.11 How to begin editing your ligand in the ICM 3D Ligand Editor. |
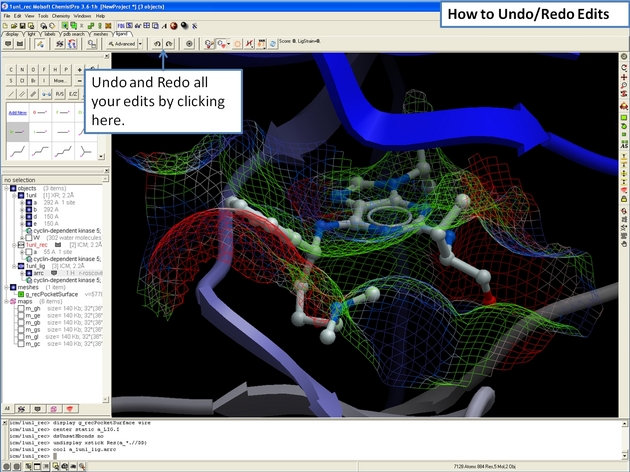
22.12.12 How to undo and redo changes in the ICM 3D Ligand Editor. |
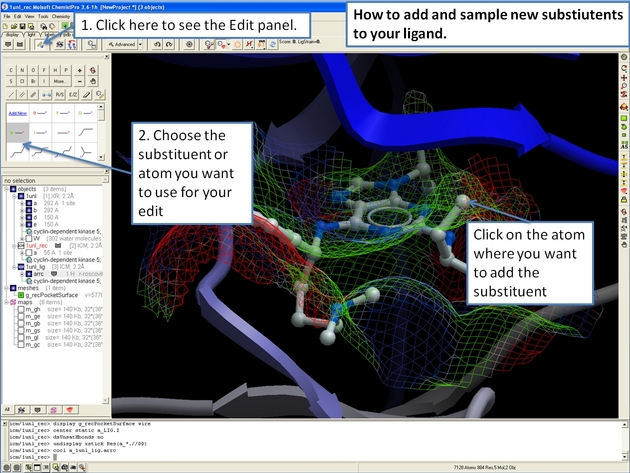
22.12.13 How to add and sample new substiutents to your ligand in the ICM 3D Ligand Editor. |
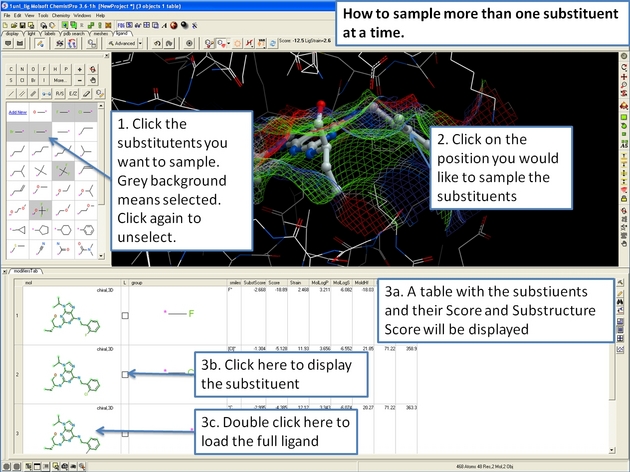
22.12.14 How to sample more than one substituent at a time in the ICM 3D Ligand Editor. |
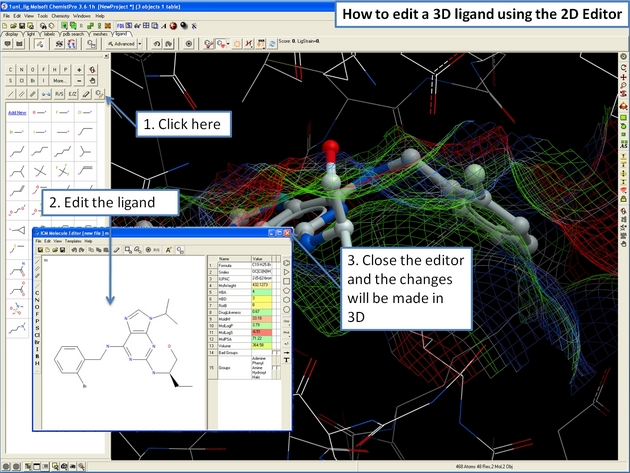
22.12.15 How to edit the ligand in 2D in the ICM 3D Ligand Editor. |
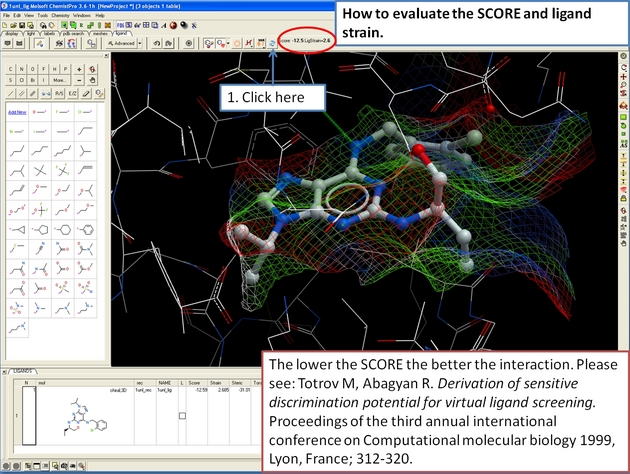
22.12.16 How to evaluate the SCORE and ligand strain.. |
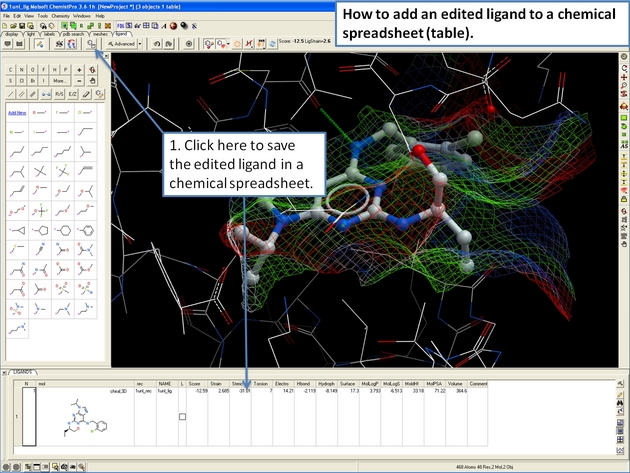
22.12.17 How to add an edited ligand to a chemical spreadsheet (table). |
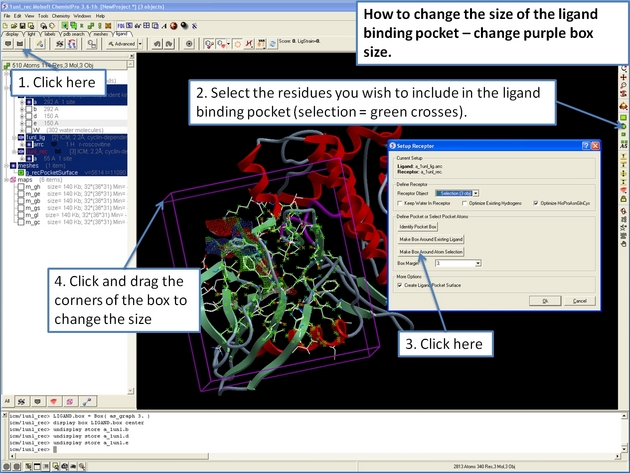
22.12.18 How to change the size of the ligand binding pocket - change purple box size. |
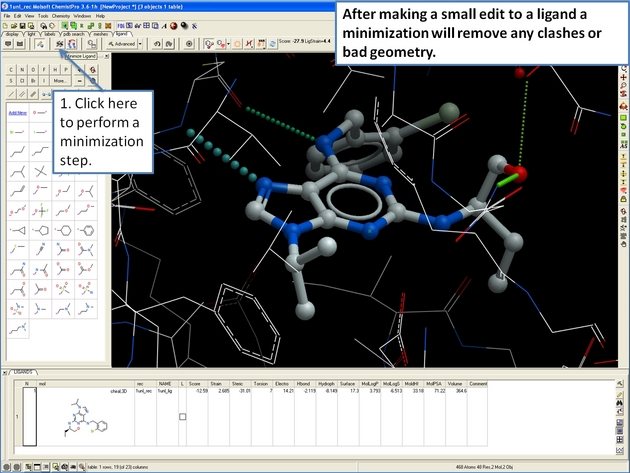
22.12.19 How to perform ligand minimization in the ICM 3D Ligand Editor. |
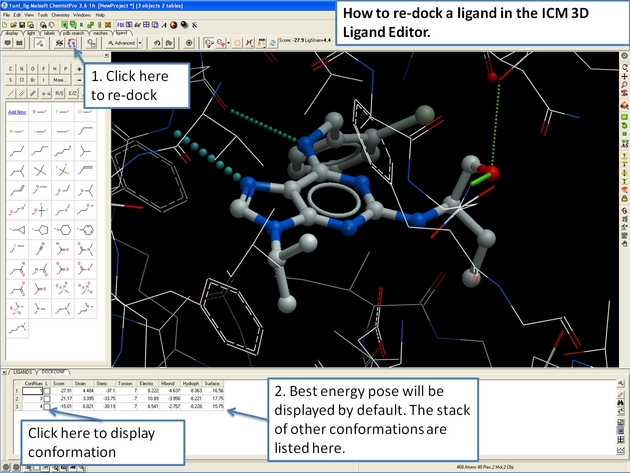
22.12.20 How to re-dock a ligand in the ICM 3D Ligand Editor. |
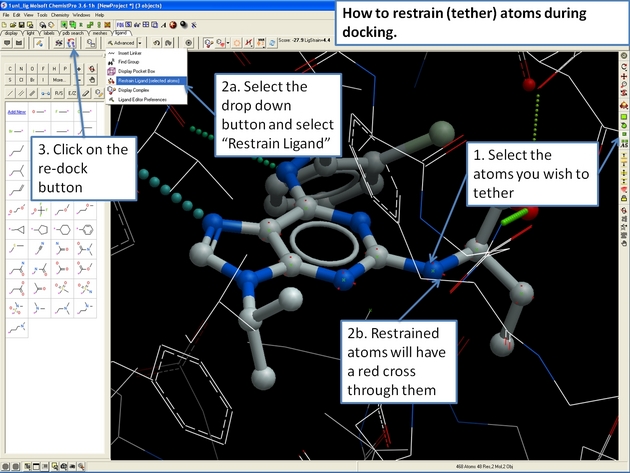
22.12.21 How to restrain (tether) atoms during docking. |
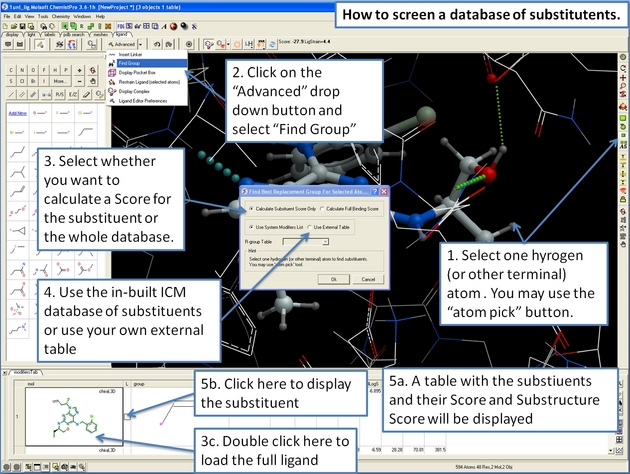
22.12.22 How to screen databases of chemical substituents. |
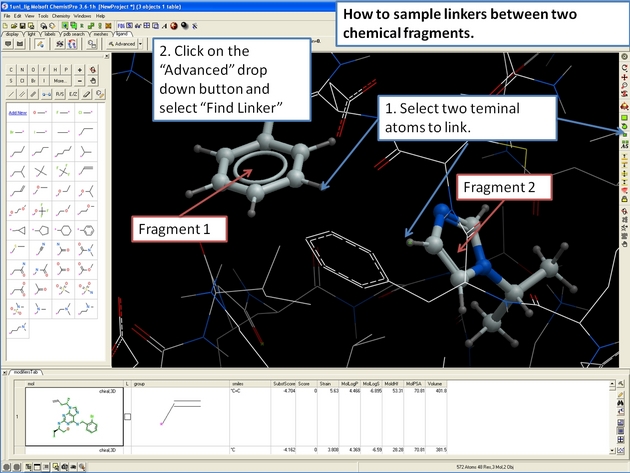
22.12.23 How to sample linkers between two chemical fragments. |
| Prev Conformer Generator | Home Up | Next Superimpose Chemicals |