| Prev | ICM User's Guide 22.10 Chemical Searching | Next |
[ Similarity | More Advanced Chemical Similarity Searching | 3D Pharmacophore Searching | 2D Pharmacophore Searching ]
22.10.1 Chemical Similarity Searching |
Objective
To find the drugs celebrex and rofecoxib in the chemical table celebrex50.sdf by performing a substructure chemical similarity search.
Background
Using ICM you can perform a compound similarity search whereby a query structure will be searched against a database of compounds. The database can be a compound database already loaded into ICM such as an SDF file or Molsoft's very own compound database called MOLCART.
Instructions
- Load the celebrex50.sdf file into ICM (File/Open). This file is provided in the ICM distribution.
- Chemistry/Chemical Search
- The ICM Molecular Editor and another menu for query search (on the right) will be displayed.
- If a molecule is already displayed in the editor you can delete it by Edit/Select All to delete
- We will start by seeing if we can identify celebrex and refecoxib from the common substructure shown below. Draw the substructure query using the Molecular Editor buttons. In this example you will draw a benzene ring with a single bond to a Sulfur atom.
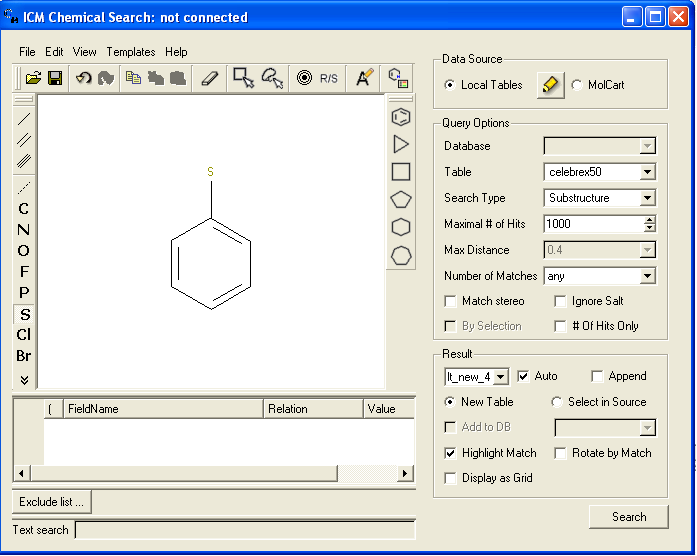
* Select the option Local Tables
* Select Celebrex50 as your database.
* Select substructure search
* Select the other options as shown in the figure above. You can experiment with different values from the drop down menu.
* Select the Search button.
* A new table will be constructed called result1 with your substructure search results contained in it. If you added Rofecoxib to the celebrex50.sdf in the previous example your results table should contain 2 hits - celebrex and rofecoxib.
<>
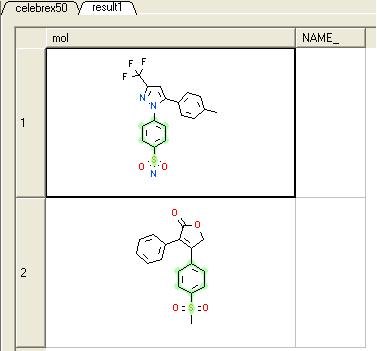
Notes and Things to Try:
- Note your substructure is highlighted in green in the results table.
- Try using the FP finger print option from the drop down Search Type button. A substructure search is a search whereby only the defined molecule in the query will be searched against the database. Whereas, a FP search which stands for fingerprint search enables any fingerprint within a structure to be searched for in the database. The "Max distance" option is available for use with the FP search and the "Matches number" option is for use with the substructure search. The option you do not require based on your search method will be blanked out. A "Max distance" value of 0 means that the search will only identify matches exactly the same as the fingerprint - the default is 0.4. The "Matches number" option allows you to stipulate how many times within a structure in the database your query can be found.
Manual References (Web Links)
Chemical Substructure/Similarity Searching
22.10.2 Advanced Chemical Similarity Searching |
Objective
To use the right click options in the chemical search window to add additional search criteria and find ways to distinguish Celebrex from Rofecoxib.
Instructions
- Load the celebrex50.sdf file into ICM (File/Open). This file is provided in the ICM distribution. Add Rofecoxib to the celebrex50.sdf file as described in the chemical-edit tutorial.
- Chemistry/Chemical Search
- The ICM Molecular Editor and another menu for query search (on the right) will be displayed.
- If a molecule is already displayed in the editor you can delete it by Edit/Select All to delete
- Follow the search instructions described in the previous example with the following chemical search substructures:
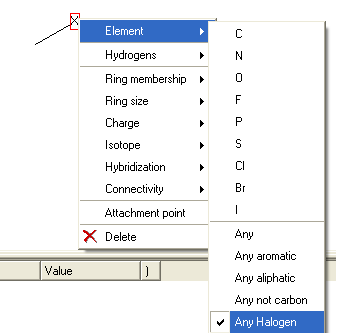
Celebrex contains halogen atoms and Rofecoxib does not - therefore one way to distinguish the two would be a simple filter as shown below.
One of the key features between Celebrex and Rofecoxib is a benzene ring connected to a five-membered ring. The difference is that in celebrex the connection point is with a nitrogen atom and in Rofecoxib the connection point is with a carbon atom. Therefore to retrieve both Celebrex and Rofecoxib in the results table you would need to right click and select Element/Any (*) and select Ring Size 5 (r5) or to retrieve only one you would need to specify nitrogen or carbon at the connection point.
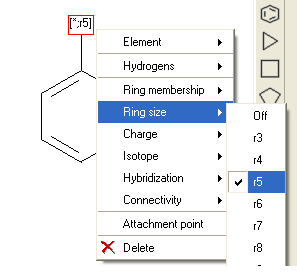
You can also perform the same query using Ring Membership (R1) or Attachment Point.
Manual References (Web Links)
22.10.3 3D Pharmacophore Searching |
Objective Undertake a 3D pharmacophore search of a table containing 3D coordinates.
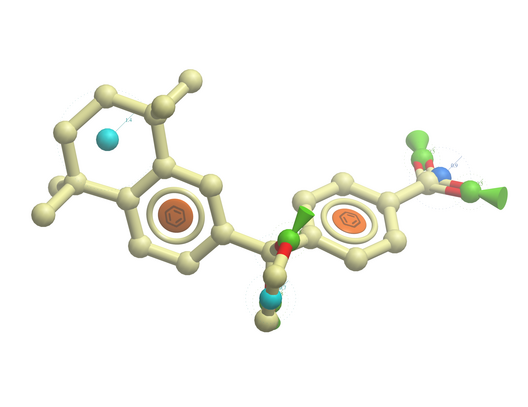
- File/Open example_ph4.icb (this file is provided in the ICM distribution and therefore can be found in $ICMHOME or in Windows Program Files/MolSoft
- In this example the 3D pharmacophore has already been extracted from a ligand. To find out how to generate a 3D pharmacophore see the section entitled Pharmacophore Draw 3D.
- In this example a table containing 3D coordinates is already provided containing 3D coordinates. The table is called t_3D.
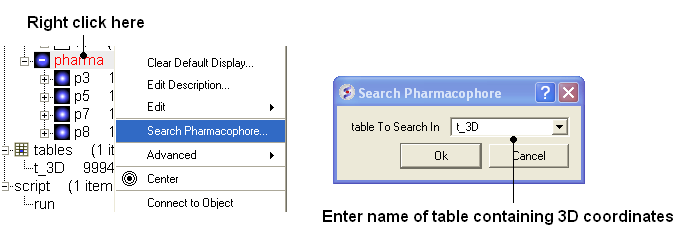
- To run 3D pharmacophore searching right click on the name of the pharmacophore object in the ICM Workspace and select Search Pharmacophore.
- Select the table t_3D from the drop down list and click OK.
- A table of search results will be displayed.
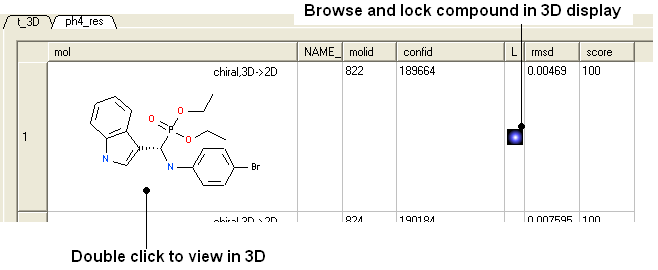
- You can browse the results by clicking on the table and the ligand will be displayed in the graphical display.
- Remember you can use the check boxes in column L to lock compounds and overlay them.
Manual References (Web Links)
22.10.4 2D Pharmacophore Searching |
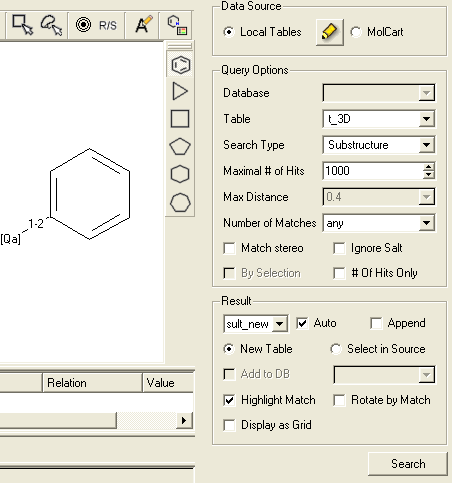
Objective Undertake a 2D pharmacophore search of a chemical spreadsheet.
- File/Open example_ph4.icb (this file is provided in the ICM distribution and therefore can be found in $ICMHOME or in Windows Program Files/MolSoft
- Chemistry/Chemical Search
- Draw the query as shown below using the
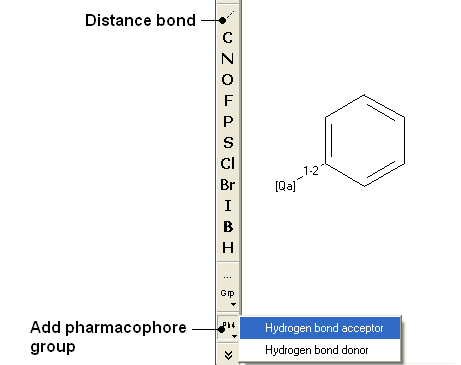
- Fill in the query and results option as shown below.
Manual References (Web Links)
| Prev Edit Chemical | Home Up | Next Chemical 2D to 3D |