| Prev | ICM User's Guide 22.2 Graphical Selections Tutorial | Next |
[ Basic Selections | Sequence Selections | Spherical Selection | Filter Selection | Residue Propogate ]
All buttons relating to graphical selections are shaded green. These buttons are shown here and are located at the top of the graphical user interface and along the side.
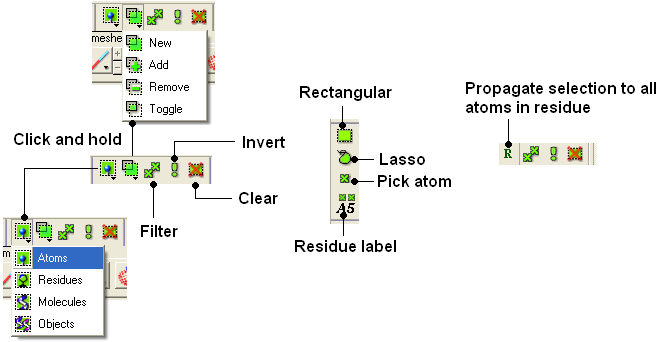
The File/New window is a great place to start learning how to use ICM as it provides a quick and easy way to generate new peptides, compounds, DNA/RNA, sequences and graphical objects.
The examples in this section will take you through the basics of making selections in the graphical user interface using objects generated from File/new.
22.2.1 Making Basic Selections |
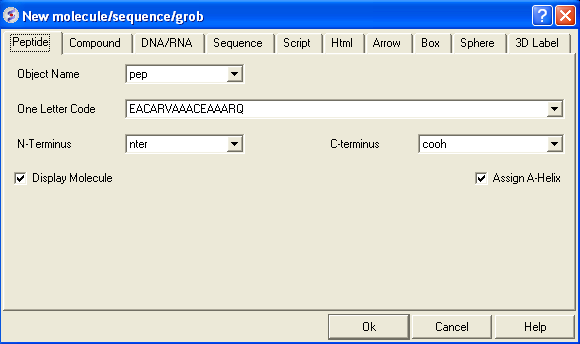
- File/New - Select Peptide Tab and use the default peptide entries and select OK. A peptide as shown below will be displayed in ribbon format and the cysteine side-chains in xstick.
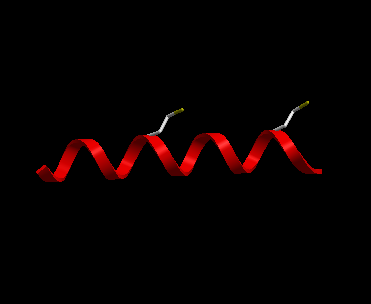
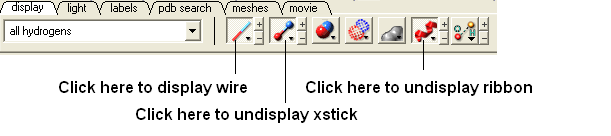
Now let us display the peptide in wire format and remove the ribbon and xstick representation
To do this:
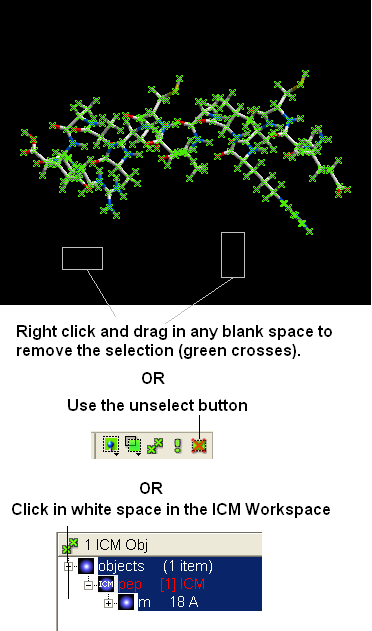
- Select the whole object by double clicking on the name of the object "pep" in the ICM Workspace. When selected it will be highlighted in blue in the ICM Workspace and green crosses in the graphical display.
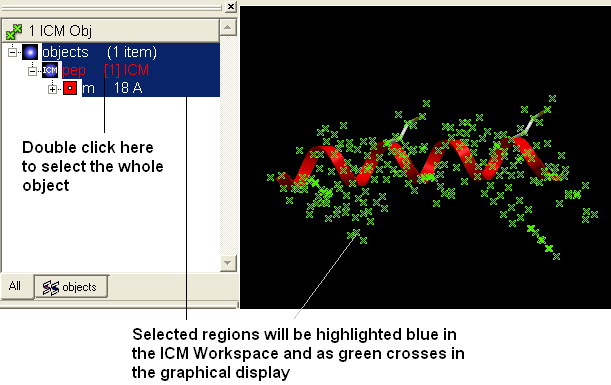
- Select the "Display Tab" and select the wire button to display wire representation. Select the ribbon button and xstick button to undisplay ribbon and xstick.
- Your peptide will still be selected. See below on how to remove selections.
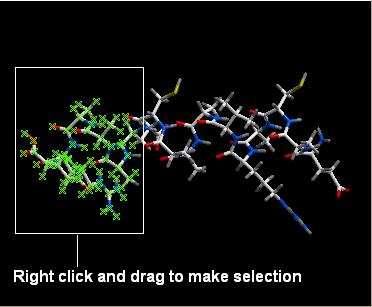
Now we will display different parts of the peptide in different representations.
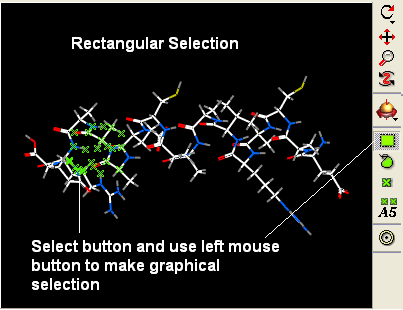
- Right click and drag over a region of the peptide you would like to change.
- Use the display panel to select the represenation you would like to display.
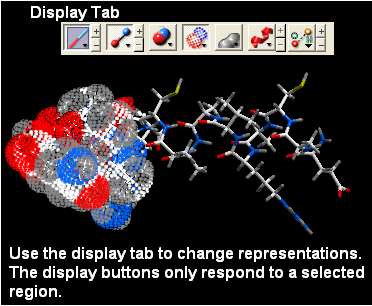
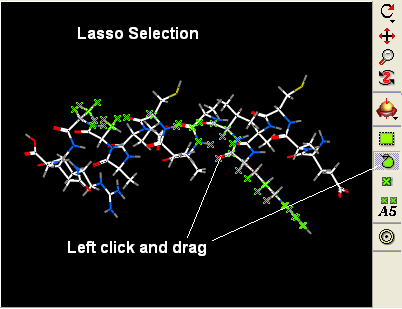
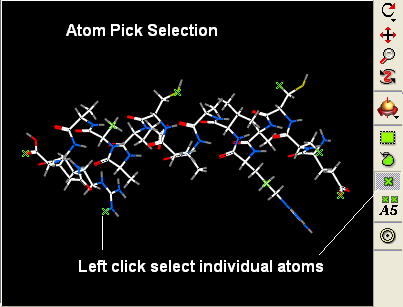
Other ways of making selections.
22.2.2 Making Sequence Selections in the ICM Workspace |
- File/New - Select Peptide Tab and use the default peptide entries and select OK. A peptide as shown below will be displayed in ribbon format.
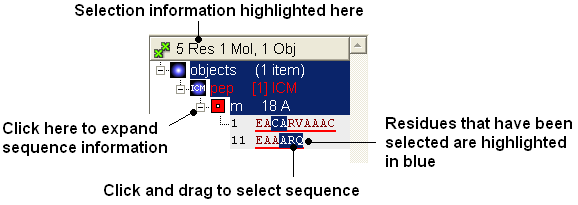
Selections can be made on the sequence of the peptide. This can be done by expanding the sequence display in the ICM Workspace and clicking and dragging on the sequence as shown below. Non-contiguous selections can be made by clicking/dragging over the sequence whilst holding down the Ctrl key.
22.2.3 Making a Spherical Selection. |
In this example we will select the residues surrounding a ligand.
- PDB Search tab (PDB code 1MVC)
- Right click on the ligand bm6 in the ICM Workspace.
- Select the Neighbors option
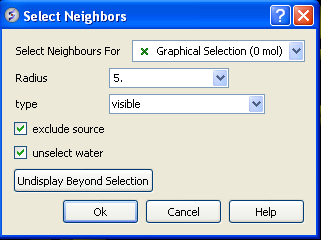
- In the "Select Neighbors For" box leave as Graphical Selection (1 mol)
- Enter Radius 5.
- Enter type same_object_other_chains
- Choose exclude source and unselect water.
- You will see green crosses surrounding the ligand binding pocket.
22.2.4 Filtering a Selection. |
In this example we will filter the selection made in the previous example and select only His, Asn, Gln and Pro residues surrounding the ligand.
- Make a spherical selection surrounding the ligand in PDB structure *1MVC (See earlier example).
- Click on the Filter graphical selection button.
- Click on the Res tab
- Select His, Asn, Gln and Pro residues.
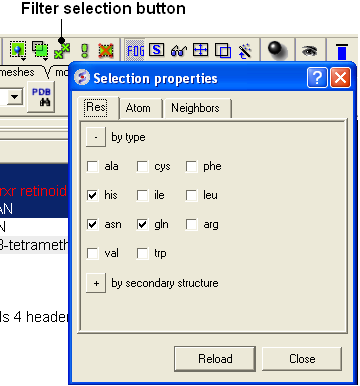
| NOTE: Only amino acid residues in the current selection will be available in the Filter. |
Display the selected residues in wire format.
- Change the selection from Atom to Residue.
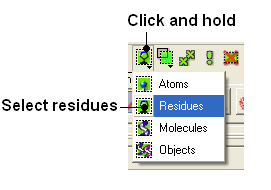
- Select the wire representation button in the display tab.
22.2.5 Propogating a selection to all atoms in a residue. |
Sometimes it is useful to propogate a selection to all atoms in a residue. For example when selecting the residues surrounding a ligand within a certain angstrom radius the selection will only pick up certain atoms of a residue. The button shown below can be used to propogate the selection to all atoms in a residue.

| Prev 2D and 3D Labels | Home Up | Next 3D Documents |